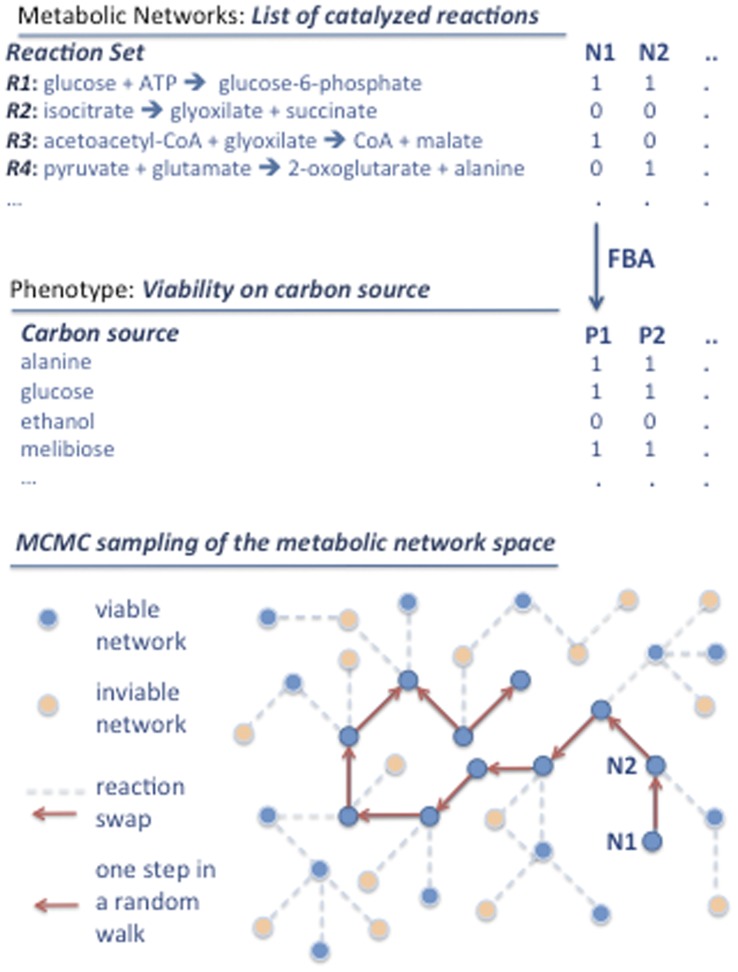
Figure 5. Exploration of a metabolic network space.
Metabolic networks can be viewed as subsets of enzyme-catalyzed metabolic reactions in a global reaction set. Formally, they can be represented as binary vectors listing the reactions catalyzed by enzymes in an organism, as indicated for two hypothetical metabolic networks (N1, N2) in the figure. Metabolic phenotypes are computed from metabolic networks using FBA. They can be represented as binary vectors indicating the carbon sources (i.e.: alanine, glucose, melibiose,…) on which a network is viable, that is, on which it can synthesize a given set of (biomass) molecules. Neighboring networks (blue circles linked by edges) differ by a single reaction swap (edges between circles) that leaves the metabolic phenotype unchanged. A reaction swap consists of two changes: one random reaction addition (R4 in the example) and one random reaction deletion (R3 in the example). A series of successful reaction swaps is called a random walk (indicated by red arrows). The Markov Chain Monte Carlo (MCMC) technique allows one to randomly sample networks with a given phenotype by generating long random walks through genotype space, where each step in a walk consists of a reaction swap. The advantage of using reaction swaps is that they leave the number of reactions constant.