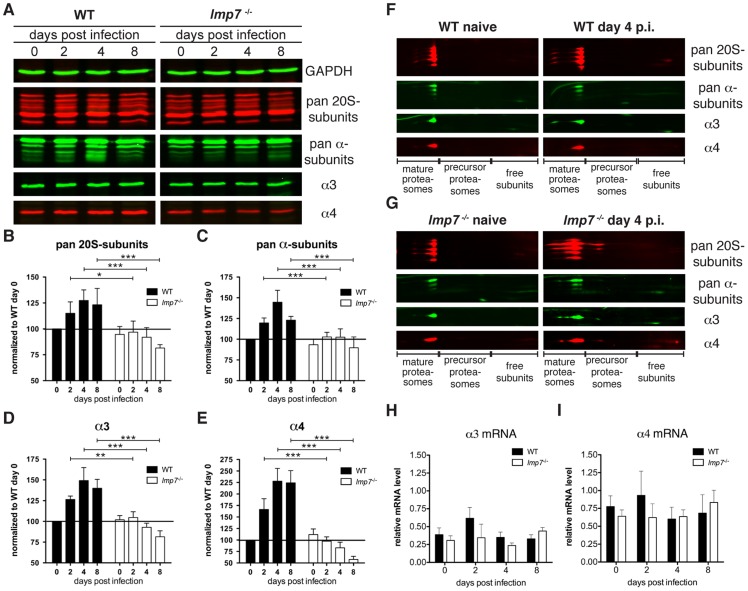
Figure 6. Relative quantification of mature proteasomes in WT and lmp7 − /− mice during the course of listeria infection.
WT and lmp7−/− mice were infected i.v. with 5×103 cfu of L. monocytogenes and livers of three to four mice per group were pooled for two-colour fluorescent immunoblot analysis. Naïve mice (day 0) were used as controls in both groups of mice. Each membrane was stained against GAPDH as loading control. Further, membranes were stained with antibodies recognizing pan-20S subunits, pan-α-subunits, α3 and α4as indicated. Representative blots of three independent experiments are shown (A). Following densitometric analysis the relative protein abundance of pan-20S subunits (B), pan-α-subunits (C), α3 (D) and α4E was expressed as band intensities normalized to WT day 0, which was calculated as follows: (band intensity of proteasome subunit X at day Y/band intensity of GAPDH at day Y)/(band intensity of proteasome subunit X in WT day 0/band intensity of GAPDH in WT day 0) × 100). The given results are means ± standard deviation of three independent infection experiments and each sample was analysed in duplicates during immunoblot analysis (B–E). To analyse in which proteasome complexes the analysed structural subunits are integrated, 2D two-colour fluorescent immunoblot analysis was performed on liver lysates of naïve and infected WT (F) and lmp7−/− mice (G). Brackets mark the areas, in which the different proteasome fractions are found. qPCR analysis on total liver cDNA was performed to assess the mRNA expression of α3 (H) and α4I during the course of listeria infection in WT and lmp7−/− mice. The relative mRNA expression of the proteasome subunits was calculated by normalization to the housekeeping gene RPS9 using the ΔΔCT method. Each value represents mean ± standard deviation of three individual mice.