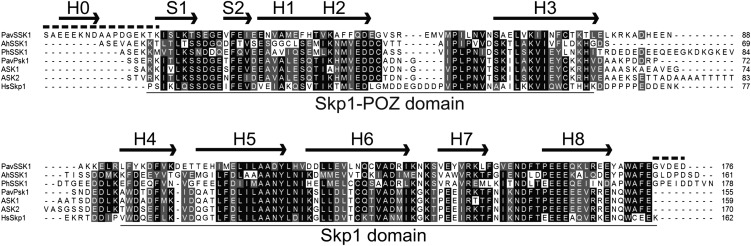
Figure 2.
Sequence alignment of SSK1s and typical Skp1-like proteins. The putative amino acid sequence of PavSSK1 was aligned with that of AhSSK1 (ABC84199) from Antirrhinum, PhSSK1 (ACT35733) from P. hybrida, ASK1 (NM_106245) and ASK2 (NM_123584) from Arabidopsis, PavPSK1 (from this study) from sweet cherry, and human HsSkp1 (P63208). The N-terminal Met residue was excluded because its excision was predicted by the TermiNator (http://www.isv.cnrs-gif.fr/terminator3/index.html; Frottin et al., 2006; Martinez et al., 2008). Black and gray blocks indicate identical and similar amino acid residues, respectively. Top arrows and dashed lines represent the secondary structure and the intrinsically disordered regions of PavSSK1, respectively. H, α-helix; S, β-sheet.