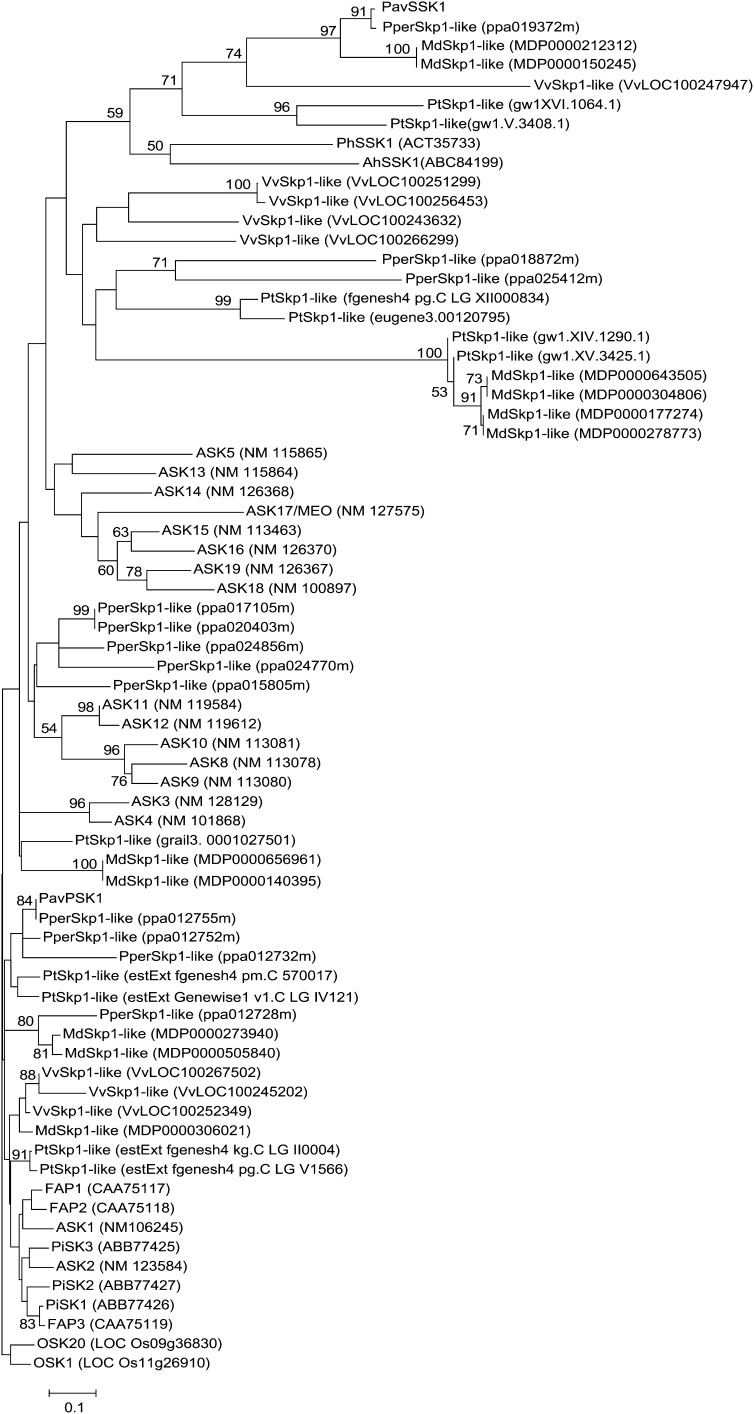
Figure 3.
NJ tree constructed using aligned deduced amino acid sequences of PavSSK1 and other plant Skp1-like proteins. Skp1-like proteins shorter than about 200 amino acid residues that possessed both Skp1 and Skp1-POZ domains were used to construct a NJ tree. The deduced amino acid sequences were from Arabidopsis (ASK1-19), grapevine (Vitis vinifera; VvSkp1-likes), poplar (Populus spp.; PtSkp1-likes), apple (Malus domestica; MdSkp1-likes), peach (PperSkp1-likes), Antirrhinum (AhSSK1 and FAP1-3), Petunia (PhSSK1 and PiSKP1-3), and rice (OSK1 and OSK20). The NJ tree was generated with 1,000 bootstrap replicates. OSK1 and OSK20, which had the best blast scores among OSKs when PavSSK1 was used as a query, were used as outgroups to construct the NJ tree.