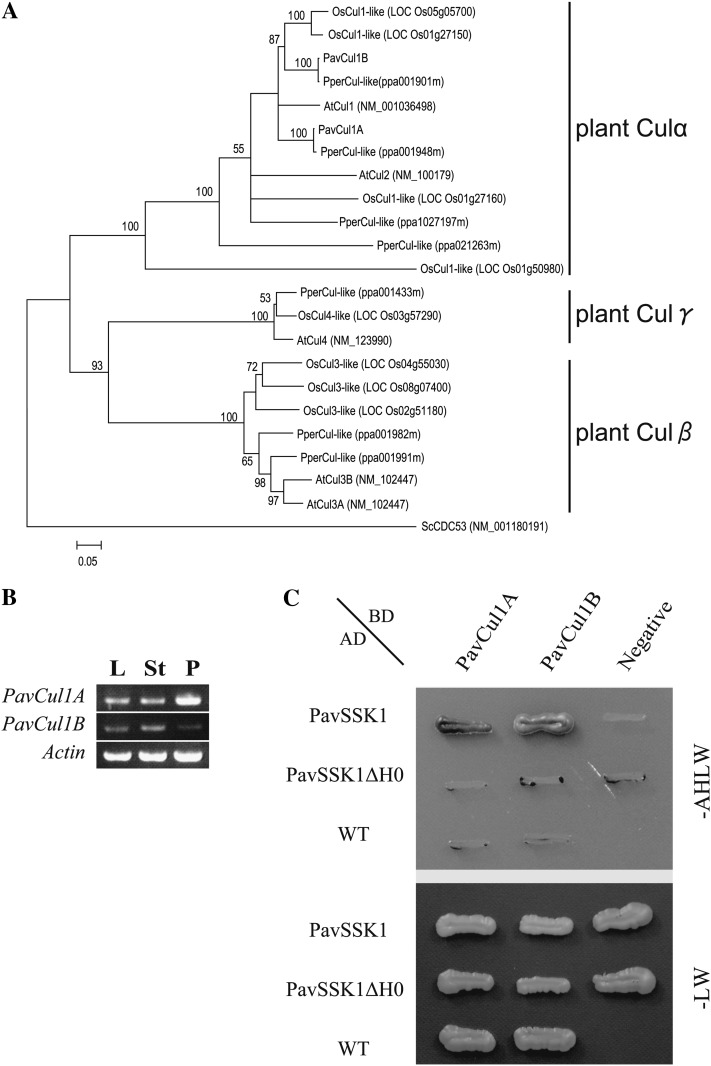
Figure 5.
Identification of the pollen-expressed sweet cherry Cul1s. A, An NJ tree was constructed based on aligned deduced amino acid sequences of Prunus Cul-likes. Deduced amino acid sequences were from peach (PperCul-likes), sweet cherry (PavCul1A and PavCul1B), Arabidopsis (AtCul1-4), and rice (OsCul1–4-likes). The NJ tree was generated with 1,000 bootstrap replicates. Budding yeast Cul α (ScCDC53) was defined as the outgroup. B, PavCul1A and PavCul1B expression profiles. Total RNAs from leaves (L), styles (St), and pollen (P) were used as a template for cDNA synthesis and RT-PCR with gene-specific primer pairs. C, Y2H analysis to investigate interactions between PavSSK and PavCul1s. A colony of AH109 cotransformants was grown on nonselective (-LW) and selective (-AHLW) media for 10 d at 25°C. The empty pAD-GAL4-2.1 vector (WT) and the empty pGBKT7 vector (Negative) were used as negative controls.