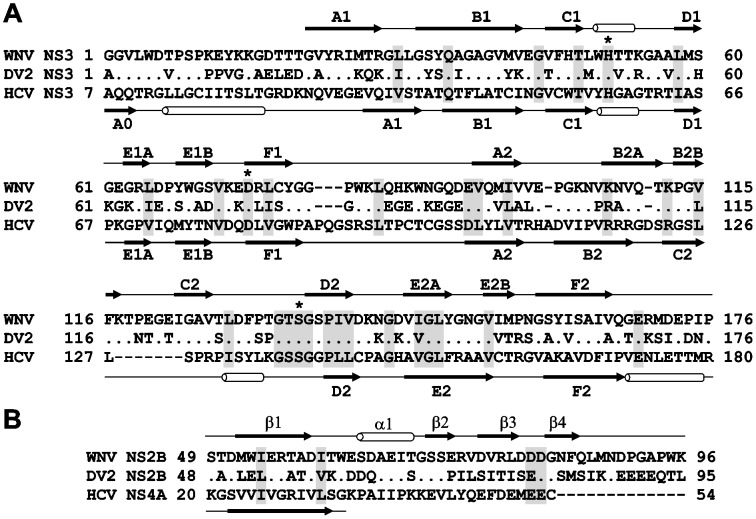
Figure 2. Structural similarity of the flaviviral NS3 proteinases.
(A) Sequence alignment of NS3 proteinases. Asterisks mark the catalytic triad. Identical and homologous residue positions are shaded gray. (B) Sequence alignment of the flaviviral WNV and DV2 NS2B and HCV NS4A (PDB 3EYD) co-factors. Dots indicate identical residues. Secondary structure elements above and below the sequences are for WNV NS2B-NS3 proteinase (PDB 2IJO) and HCV NS3/4A (PDB 3EYD), respectively. The secondary structure of the minimal, 14-residue, NS4A co-factor required for activation of the NS3 proteinase in vitro is shown. WNV, West Nile virus. DV2, Dengue virus type 2.