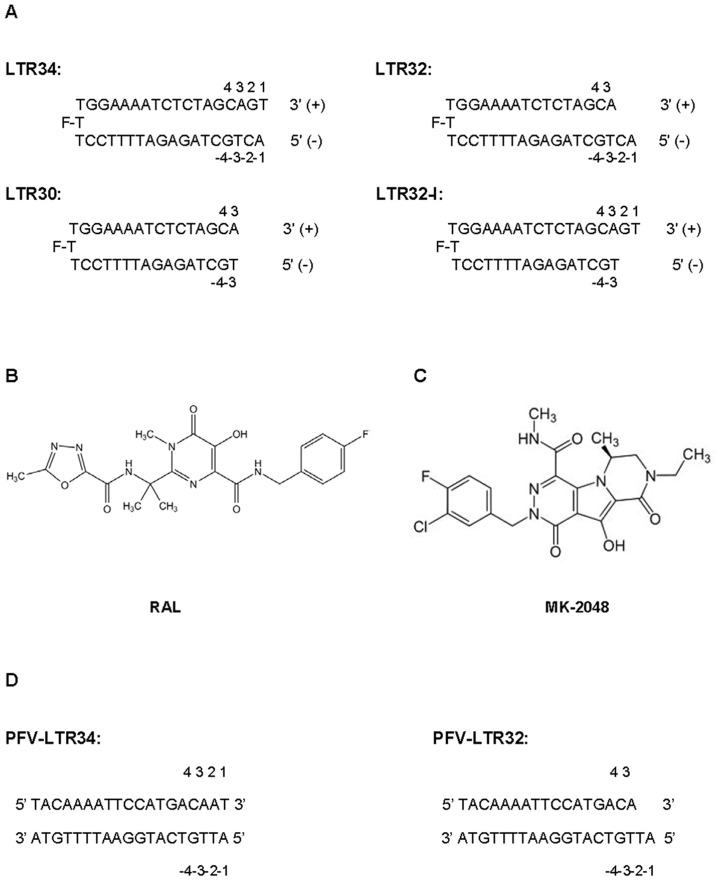
Figure 1. Molecules used in this study.
(A) The oligonucleotides are designed to adopt a hairpin structure, folded around a three thymine loop whose central thymine bears the fluorescein reporter. LTR34: unprocessed U5-LTR end with a 17 base pair stem. The numbering of the four last base pairs (+1 to +4 and −1 to −4) starts from the ultimate 3′nucleotide on the upper (+) strand and from the ultimate 5′ nucleotide on the lower (−) strand; LTR32, processed U5-LTR, with 5′A−1C−23′ as overhang on the (−) strand; LTR32-I, inversed LTR32, with 5′G2T13′ as overhang in the (+) strand; and LTR30, doubly deleted LTR34 (blunt-ended DNA). (B) Chemical structure of the here studied RAL (MK-0518). (C) Chemical structure of MK-2048. This compound is given as an example of INSTI of second generation, inducing less resistance mutations in IN. (D) PFV LTR sequences used in calculations. The numbering of the four last base pairs (+1 to +4 and −1 to −4) is the same as in (A).