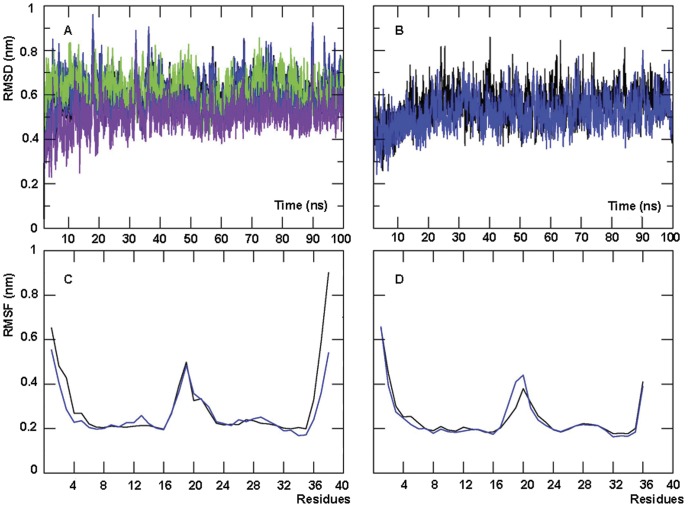
Figure 6. MD simulations of the RAL-LTR34 and RAL-LTR32 complex systems (PFV oligonucleotides), using GROMACS with the AMBER force field.
(A) Time evolution of RMSD (root mean square deviation) values based on all the heavy atoms for the two LTR34 trajectories (black: LTR34-1 and blue: LTR34-2). RMSD calculations for a single trajectory were also performed using the sugar C4′ atoms (green: LTR34-1) and repeated for LTR34 devoid of 3′-AT (purple). (B) Time evolution of RMSD values of LTR32 for two trajectories (black: LTR32-1 and blue: LTR32-2). (C) RMSF (root mean square fluctuation) variations of sugar C4′ atoms for LTR34 and (D) RMSF variations of sugar C4′ atoms for LTR32.