Figure 4.
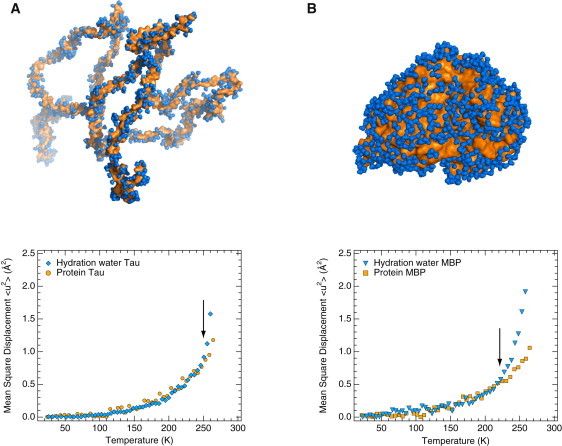
Dynamical coupling of protein dynamics with hydration-water motions for different protein classes. Temperature-dependent atomic MSDs of proteins (orange data points) are compared with those of hydration water (blue data points) and are identical to the ones shown in Fig. 1. (A) IDP tau. (B) Folded, globular MBP (7). An ensemble of 100 tau structures was generated using a coil library (58) and a structure with a calculated radius of gyration (63 Å) identical to that measured by SAXS (63 Å) was chosen for the upper half of panel A. Approximately 1000 water molecules forming the first hydration shell are shown in panel A, corresponding to the experimental hydration level of 0.4 g water/g protein. Data were obtained on the IN16 spectrometer at the ILL, Grenoble, France. Arrows indicate the temperature at which water and protein MSD diverge (250 K (A) and 220 K (B) for tau and MBP, respectively). MSDs > 260 K were omitted for the protein (tau and MBP) experiments for the sake of comparison with hydration-water MSDs (tau and MBP), which could be determined only up to 260 K (see Materials and Methods section).
