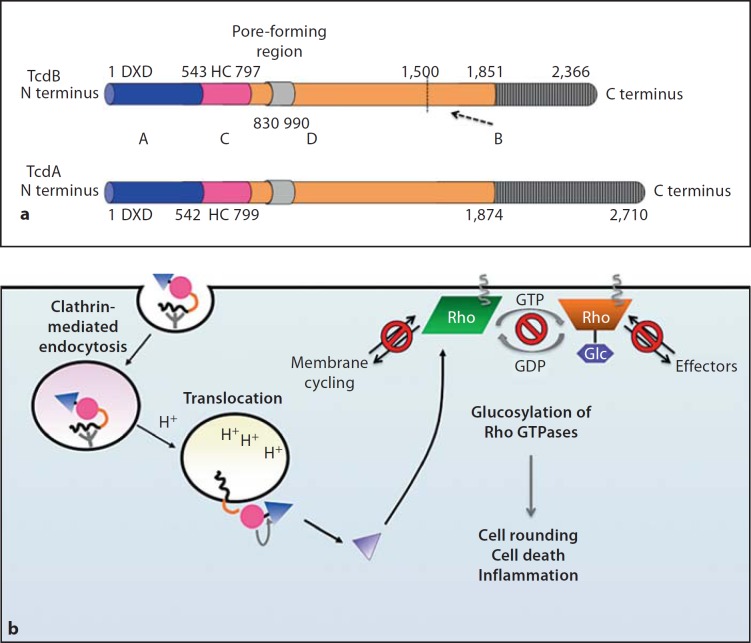
Fig. 2.
aABCD structure of glucosylating toxins TcdA and TcdB. Functional and structural domain boundaries are marked, with active site residues being indicated. A = ‘Activity’ domain of Glc (blue); C = ‘cutting’ domain/autoprocessing CPD (pink); D = ‘delivery’ domain/translocation domain (orange); B = ‘binding’ domain/receptor binding domain (grey). For the D domain, the minimal pore-forming region consists of residues 830–990, while residues 1–1,850 are sufficient to mediate intoxication of target cells. The B domain contains multiple repeat sequences (CROPs), which range in size from 21–50 residues and are repeated throughout the C-terminus of the protein. The CROPs are more divergent and less frequent in TcdB than in TcdA. The dashed arrow indicates a putative minor receptor binding domain (residues 1,500–1,850) identified by deletion analyses [38, 40]. b Intoxication of target cells by TcdA and TcdB. Binding of the B domain to unknown receptors on target cells results in clathrin-dependent receptor-mediated endocytosis. During acidification of the endosome, conformational changes occur within the toxin that result in pore formation by the D domain and translocation of the A domain into the cytosol. Exposure of the C domain to InsP6 (yellow) activates its protease function, resulting in toxin autoprocessing and release of the A domain into the cytosol. The A domain glucosylates Rho GTPases (Rho, Rac, Cdc42 family) on a conserved Thr residue, which prevents Rho GTPases from interacting with their cognate effectors and sequesters them at the membrane.