Figure 7.
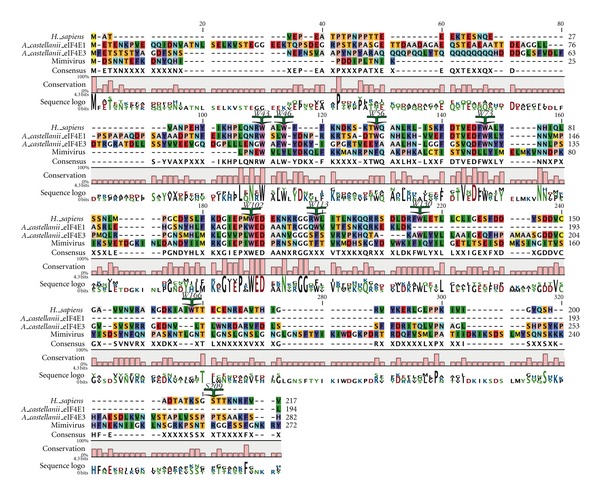
Comparison of the sequence of mimivirus eIF4E with those of an Acanthamoeba species. Alignment of the amino acid sequences of mimivirus eIF4E with eIF4Es from its host Acanthamoeba castellani. Amino acid sequences were aligned with T-coffee using the BLOSUM62MT scoring matrix in CLC Main Workbench. A. castellani sequences were derived from the Protist EST Program (PEP) in advance of scientific publication and acceptance by GenBank at http://amoebidia.bcm.umontreal.ca/public/pepdb/agrm.php. This site is no longer publically available. To facilitate comparison of the sequences, the residues conserved in Class I eIF4Es in multicellular organisms are indicated and numbered as in human eIF4E-1: W43, W46, W56, W73, W102, W113, W130, W166, and S209.
