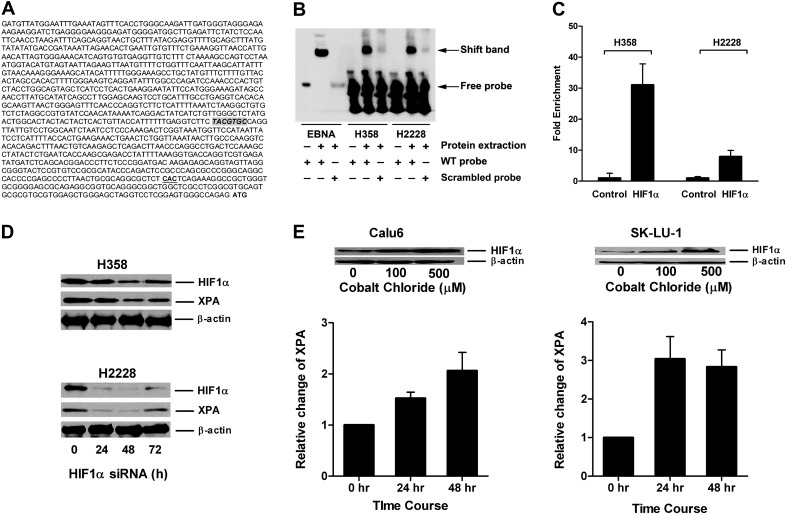
Fig. 1.
HIF1α regulated expression of XPA in lung cancer cell lines. (A) Location of HRE-binding site TACGTGC in XPA promoter relative to the transcriptional start site (CAC) and the translational start site (ATG) is shown. (B) The binding of HIF1α to HRE is demonstrated by EMSA assay. Whole cell extracts from H358 and H2228 were incubated with HRE-containing probe and then subjected to EMSA. Shift band is shown compared with the positive control (Epstein-Barr Nuclear Antigen, EBNA) in the presence of protein extract and consensus oligonucleotide for HIF1α binding (wild-type probe) compared with the scrambled probe. (C) Binding of HIF1α protein to the XPA promoter. Antibody to HIF1α was used to assess binding of this protein to the XPA promoter using EZ ChIP™. The fold enrichment of HIF1α in XPA promoter in H358 and H2228 compared with normal mouse IgG is shown. (D) Inhibition of HIF1α by small interfering RNA (siRNA) reduces expression of XPA in lung cancer cell lines. Expression levels determined by western blotting of HIF1α and XPA following HIF1α transient knockdown in H358 and H2228 are shown at indicated time points posttransfection. (E) Cobalt chloride treatment increases expression of HIF1α and XPA in lung cancer cell lines. Calu6 and SK-LU-1 cells were treated up to 48 h with 0, 100 or 500 μM cobalt chloride. Dose response increase in expression of HIF1α is shown along with increased expression of XPA seen with the 500 μM dose.