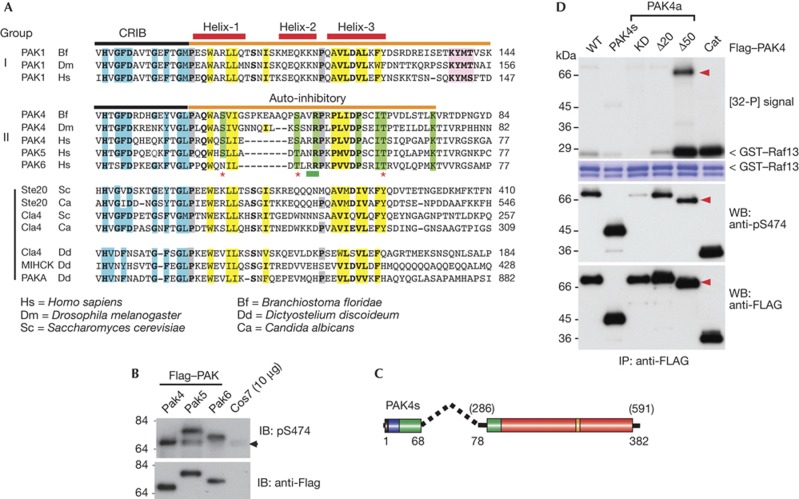
Figure 2.
Identification of a conserved PAK4 auto-inhibitory domain (AID). (A) An alignment of the AID (orange bar) comparing group I PAKs with those from metazoan PAK4 and fungal and protozoan PAKs. The three α-helices in the PAK1 AID are marked (red bars). The A-loop binding motif (KYMS/T, pink box) is not found in fungal or protozoan PAKs. Proline residues preceding helixes 1 and 3 are marked in grey. Conserved residues involved in Cdc42 binding are in blue. Hydrophobic AID residues are highlighted in yellow, while those conserved only among group II PAKs are in green. The position of the PAK4 AID mutations tested in this study are indicated by asterisk marks (potential phosphorylation sites) or underlined in green (R48, R49). (B) Flag–tagged PAK4/5 and 6 expressed in COS7 were analysed for activation loop phosphorylation. (C) Schematic of a minimal synthetic construct PAK4(s) comprising residues PAK4(1–68) joined to PAK4(286–591) with a 10-residue linker. (D) The various Flag–PAK4 were used for in vitro kinase assays performed as described in Fig 1. The A-loop phosphorylation was probed using CS#3241. Cat, PAK4(286–591); CRIB, Cdc42–Rac interaction/binding; GST, glutathione S-transferase; IP, immunoprecipitated; KD, kinase dead, PAK, p21-activated kinase; PAK4a(K350M); WB, western blot; WT, wild type; Δ20, PAK4a(21–591) and Δ50, PAK4a(51–591).