Table 2. Thermodynamic parameters for the unfolding of T. aquaticus SCS.
| Protein |
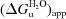 † (kJmol1)
† (kJmol1) |
m † (kJmol1 M 1) | Correlation coefficient† | [GdmCl]1/2 ‡ (M) |
|---|---|---|---|---|
| Whole enzyme | ||||
| First transition | 29 4 | 13 2 | 0.986 | 2.2 |
| Second transition | 25 1 | 7.1 0.4 | 0.998 | 3.7 |
| -Subunit | 6.7 0.4 | 3.1 0.2 | 0.998 | 2.2 |
| -Subunit | 10.0 0.8 | 5.4 0.4 | 0.997 | 2.0 |
These values were calculated by extrapolation of a replot of the molar ellipticities using the equation G = RTlnK, where K = (N
obs)/(obs
U), N is the molar ellipticity observed at 0M GdmCl, U is the molar ellipticity observed at the maximum concentration of GdmCl and obs is the observed molar ellipticity. In the vicinity of the transition the resulting plot was fitted to the equation G = 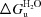 m[GdmCl] (for a review, see Myers et al., 1995 ▶) with the correlation coefficient listed.
m[GdmCl] (for a review, see Myers et al., 1995 ▶) with the correlation coefficient listed.
These values were obtained by interpolation from the graphs and are consistent with the values obtained by the calculation [GdmCl]1/2 = [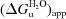 /m.
/m.
