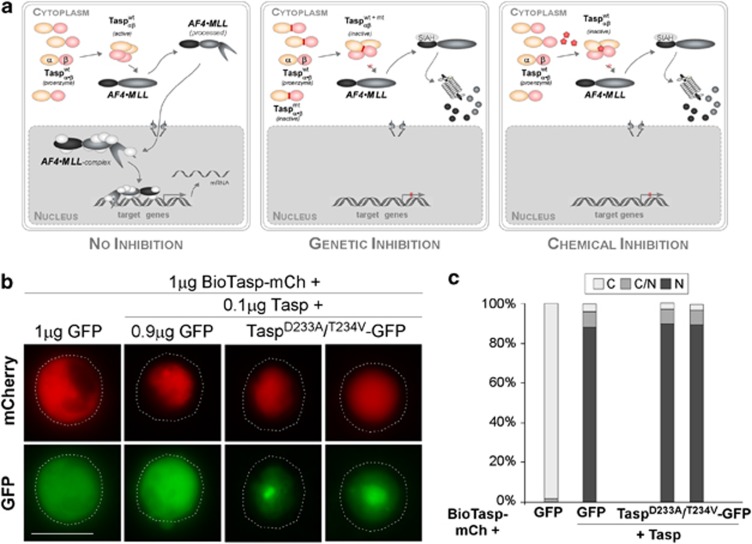
Figure 1.
(a) Strategies targeting Taspase1's oncogenic activity. Autoproteolysis of the Taspase1 proenzyme triggers formation of the active αββα heterodimer, hydrolyzing the AF4•MLL fusion protein and driving oncogenesis (left). Inhibition by overexpression of trans-dominant Taspase1 mutants results in the formation of inactive heterodimers, precluding AF4•MLL processing, and the activation of oncogenic programs (middle). Chemical Taspase1 inhibitors affect its proteolytic activity, preventing AF4•MLL processing and activation of pathological pathways (right). (b) Catalytically inactive Taspase1 mutants are not inhibitory. K562 cells were transfected with 1 μg of red fluorescent BioTaspR and 0.1 μg of Tasp-BFP, together with the indicated amounts of inactive Taspase1-green fluorescent protein (GFP) mutants or GFP expression plasmid. Even co-transfection of a ninefold excess of plasmids encoding the inactive Taspase1 variants did not affect BioTaspR processing. Localization was analyzed 48 h post transfection. GFP/mCherry (mCh) were visualized by fluorescence microscopy. Dashed lines mark cytoplasmic cell boundaries. (c) The number of cells showing cytoplasmic (C), cytoplasmic and nuclear (C/N), or nuclear (N) fluorescence was counted in at least 200 BioTaspR-expressing cells. Whereas the number of cells displaying cytoplasmic fluorescence significantly decreased by co-transfection of 0.1 μg Tasp-BFP expression plasmid, no significant trans-dominant-negative effect was evident for Taspase1 mutants.