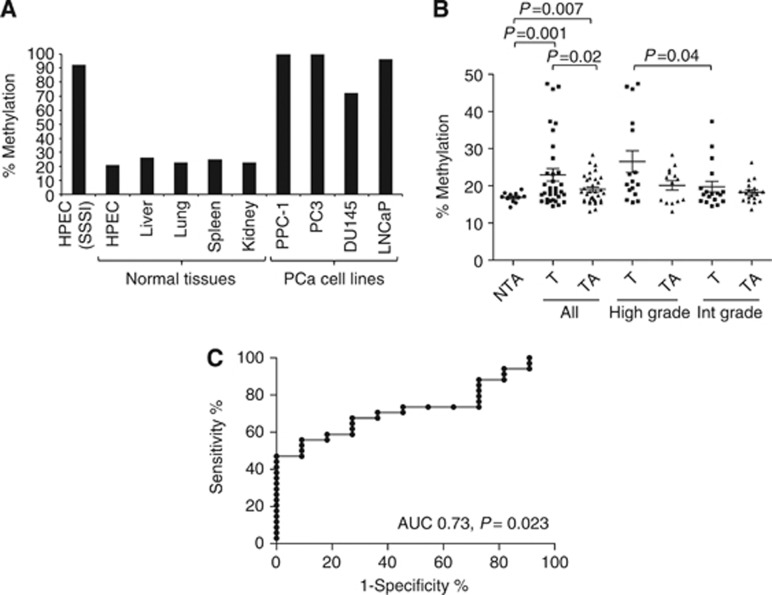
Figure 3.
Quantitative MethyLight assessment of methylation in normal tissues, human prostate epithelial cells, prostate cancer cell lines and microdissected prostate tissue specimens. Bisulphite-treated DNA was analysed using PCR and binding of fluorescently labelled MethyLight probes distinguishing methylated and unmethylated CpGs. (A) Prostate cancer cell lines PPC-1, DU145, PC3 and LNCaP demonstrated high levels of methylation (100.0%, 72.1%, 100.0% and 96%, respectively), whereas normal HPEC, liver, lung, spleen and kidney demonstrated low levels of methylation (26.3%, 22.8%, 22.8% and 24.9%, respectively). HPEC DNA treated with a methylating agent (SSSI methylase) was used as a positive control. (B) Tumour specimens (T) demonstrated increased methylation compared with matched benign tumour-associated (TA) tissue, as well as normal peripheral prostate tissue from prostates not containing cancer (NTA; P=0.02 and P=0.001, respectively). When stratified according to grade, high-grade tumours (Gleason ⩾8) demonstrated increased methylation compared with intermediate-grade tumours (Gleason ⩽7; P=0.04). (C) A receiver operating characteristic (ROC) curve was constructed to determine the frequency of hypermethylation in tumour specimens (AUC 0.74, P=0.023). The optimum cutoff of 18.2% yielded a 55.9% sensitivity and 90.9% specificity for differentiating tumour from normal specimens, and was used to define hypermethylation. Using 18.2% as a cutoff, 20/35 (57%) of tumours were hypermethylated, while only 1/11 (9%) NTA tissues were hypermethylated (P=0.006).