Fig. 1.
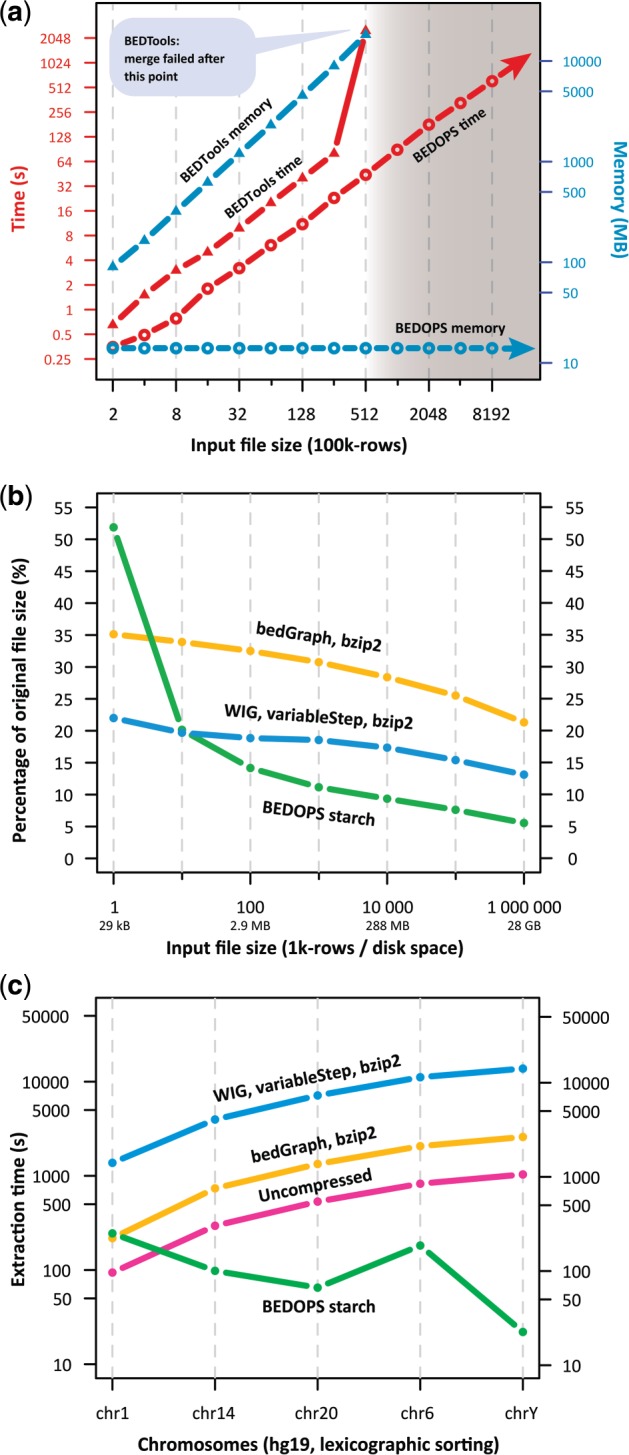
Performance results using subsets of 46-way phyloP (Pollard et al., 2010) human conservation data as inputs. (a) The time and memory resources used to merge overlapping and adjoining genomic segments as a function of input size. (b) Compressed file sizes on disk as a percentage of original uncompressed sizes. (c) Single-chromosome data extraction times from files containing >2.8×109 sorted records. BEDOPS accessed target chromosome data only, whereas conventional approaches processed inputs sequentially
