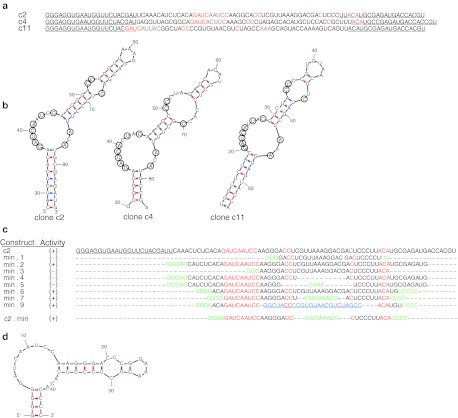
Figure 2.
Comparison of anti-TfR binding clones c2, c4, and c11. (a) Sequences of clones c2, c4, and c11 with constant regions (underlined). The additional 3′ constant region used for hybridization of a fluorescent probe for flow cytometry is not shown. Invariant residues shared between the clones are colored red. (b) Functional clones share a common fold. mfold predicted structure of c2, c4, and c11. Conserved residues highlighted in red (a) have been circled. The 5′ and 3′ ends of the sequences have been removed for clarity. (c) Minimization of c2. Truncations were made by runoff transcription from the corresponding dsDNA templates. All sequences share an additional 3′ constant region used for hybridization to a fluorescent probe for flow cytometry (data not shown). The conserved regions are highlighted in red. Residues added to start transcription, force pairing or introduce a tetraloop are indicated in green. The stem loop taken from c11 is indicated in blue. (d) mfold predicted structure of minimized anti-TfR aptamer c2.min. TfR, transferrin receptor.