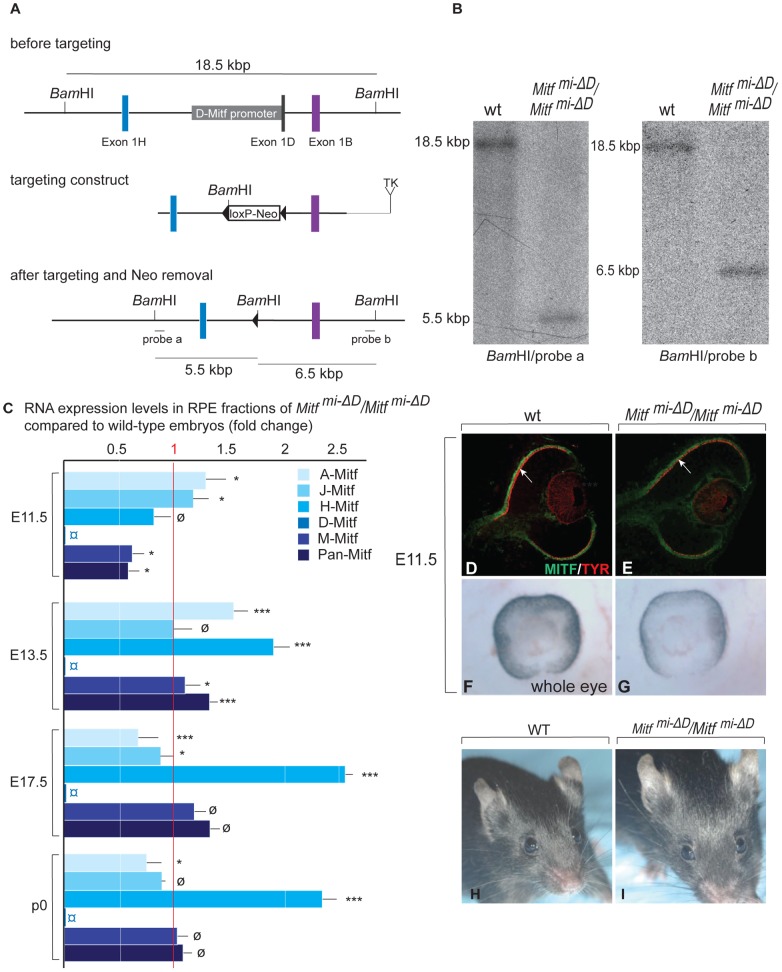
Figure 2. Generation and analysis of mice lacking the RPE–specific D-isoform of Mitf (Mitfmi-ΔD/Mitfmi-ΔD).
(A) Top: Schematic diagrams showing a region of the Mitf gene containing exons 1H, 1D, and 1B; Middle: the targeting construct with a novel BamHI restriction site and a floxed Neomycin cassette in place of 5.8 kbp of the D-Mitf promoter/exon 1D region; Bottom: Mitf gene portion after targeting. Probe ‘a’ recognizes a 5.5 kbp and probe ‘b’ a 6.5 kbp BamHI restriction fragment after targeting while both probes recognize the same 18.5 kbp fragment before targeting (see Materials and Methods for the details on construct design). (B) Southern hybridization of BamHI-restricted genomic DNA from wild-type and homozygous mutants shows the expected bands. (C) Mitf isoforms are upregulated in the RPE of Mitfmi-ΔD/Mitfmi-ΔD mice. Quantitative RT-PCR analysis for Mitf-isoforms from wild-type and Mitfmi-ΔD/Mitfmi-ΔD RPE fractions. Primers specific for Mitf isoforms A, J, H, D, and M were used to measure the respective RNAs. All values are normalized using the housekeeping gene Usf1. Results (mean values, S.D. and statistical significance [see Experimental Procedures] based on 3 biologically independent samples) are shown as fold change in RNA expression level in Mitfmi-ΔD/Mitfmi-ΔD compared to wild type. (D–G) Mice lacking the D-isoform of Mitf have reduced/delayed RPE pigmentation. Eye sections (D,E) from E11.5 wild type and Mitfmi-ΔD/Mitfmi-ΔD embryos show reduced MITF (green) and TYROSINASE (red) staining in the RPE (arrows in D,E). (F,G) Whole eye pictures show a mild reduction in pigmentation (F,G). (H,I) Adult Mitfmi-ΔD/Mitfmi-ΔD mice are indistinguishable on visual inspection from wild-type mice.