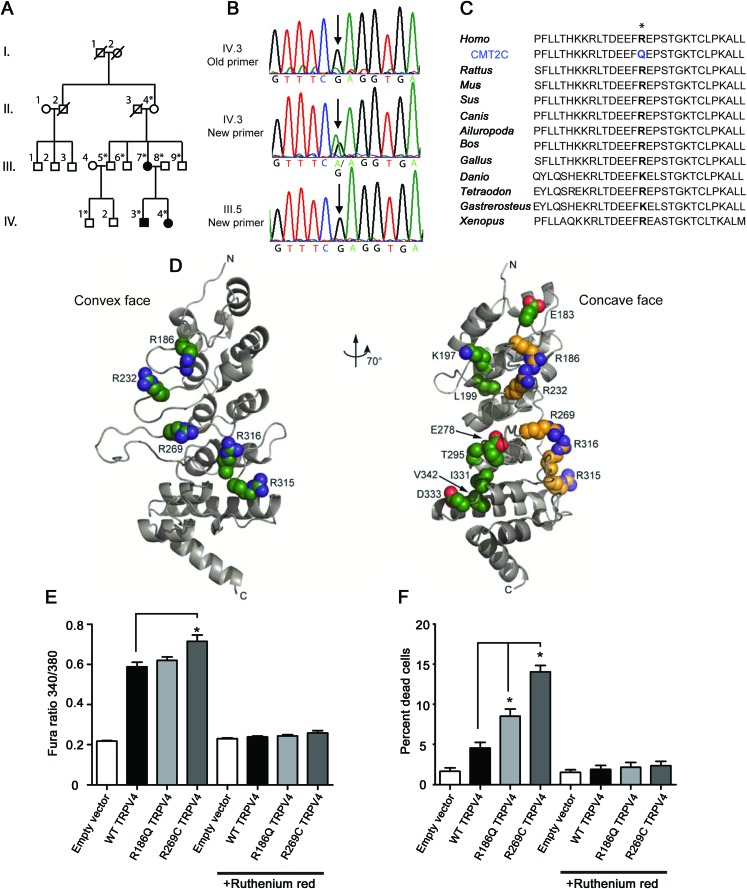
Figure 1. A family with CMT2C has a novel mutation in TRPV4.
(A) Pedigree of the family shows vertical transmission of the disease (white = unaffected, black = affected, *DNA collected). (B) Chromatographs show the c.557G>A mutation initially missed by Sanger sequencing and later identified by exome sequencing in an affected individual. This sequence variant was not apparent using the original primers (top; the “A” variant peak is comparable to the background noise), but became clearly visible when primers that do not contain single nucleotide polymorphisms (SNPs) were used to amplify the genomic DNA (middle). The sequence variant is not present in unaffected individuals (bottom). (C) The R186 residue in TRPV4 is highly conserved across a wide range of species. (D) Like previously described neuropathy-causing TRPV4 mutations, the R186 residue is situated on the convex face of the TRPV4 ankyrin repeat domain (ARD). In contrast, those skeletal dysplasia mutations occurring in the TRPV4 ARD localize to its concave face (at left: green = neuropathy mutations; at right: yellow = neuropathy mutations, green = skeletal dysplasia mutations). (E) Basal calcium levels are modestly increased in HEK293 cells expressing R186Q-TRPV4 and significantly increased in cells expressing R269C-TRPV4, compared to cells expressing WT-TRPV4 for 24 hours. The TRP channel antagonist ruthenium red completely blocks this calcium influx (*p < 0.05, n = 8 per group). (F) Approximately 10% of cells expressing R186Q-TRPV4 and 15% of cells expressing R269C-TRPV4 compared to approximately 5% of cells expressing WT-TRPV4 were dead at 36 hours post-transfection (*p < 0.01, n = 3 per group). Treatment with ruthenium red completely blocked cellular toxicity. Cells were incubated with 2 μM calcein AM and 4 μM EthD-1 for 30 minutes at room temperature and the percentage of dead cells calculated using a Zeiss AxioImager Z1 microscope and AxioVision 4.6 software.