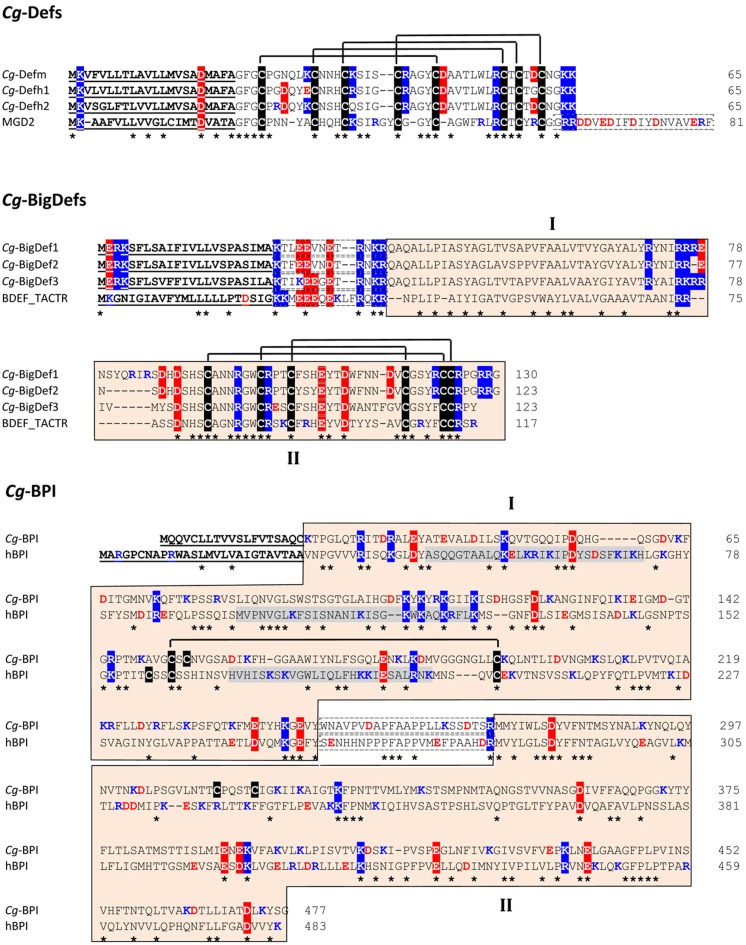
Figure 2.
Amino acid alignment of oyster antimicrobial precursors with similar proteins/polypeptides from other species. Cg-Def precursors are composed of a 22-residue signal peptide followed by a 43-residue mature peptide. By comparison, the Mytilus galloprovincialis MGD2 (GenBank AAD45118.1) carries an additional anionic proregion at C-terminal position. The amino acid sequences displayed here for Cg-Defm, Cg-Defh1, and Cg-Defh2 correspond to GenBank AJ565499, ACQ73009, and ACQ76280, respectively. The cysteine array is based on the 3D structure of Cg-Defm (PDB: 2B68). Cg-BigDef precursors are composed of a predicted signal peptide (23 residues) followed by 13-residue anionic proregion and the putative mature BigDefs (87–94 residues). Mature BigDefs are multi-domain polypeptides composed of a hydrophobic N-terminal domain (I) and a C-terminal cysteine-rich domain (β-defensin-like domain, II) shown here as orange boxes. A putative cleavage motif (RXKR) for furin-like enzymes separates both domains. Sequences of big defensins from Crassostrea gigas (Cg-BigDef1: GenBank AEE92768, Cg-BigDef2: GenBank AEE92775, Cg-BigDef3: GenBank AEE92778) are aligned here with Tachypleus tridentatus big defensin (BDEF_TACTR, GenBank: P80957). The cysteine array is based on the 3D structure of the horseshoe crab big defensin (PDB: 2RNG). Cg-BPI precursor is composed of a 19-residue signal peptide followed by a 458 amino acid protein. Its amino acid sequence (GenBank AY165040) is aligned here with human BPI (hBPI) sequence (GenBank J04739). The N-terminal (domain I) and C-terminal (domain II) barrel type domains characterized for hBPI as well as the corresponding sequences in Cg-BPI are in orange boxes (domains I and II, respectively). The proline-rich central domain is boxed with a dashed line. The three functional regions of hBPI, which display LPS-binding activity, are highlighted in gray. The conserved disulfide bridge characterized in hBPI 3D structure (PDB: 1BP1) is displayed. Amino acids are numbered on the right. Signal peptides are underlined. Cysteines are highlighted in black and disulfide bridges are displayed as black lines. Conserved residues are shown by an asterisk. Lys/Arg residues are in blue. Asp/Glu residues are in red. Positively and negatively charged residues conserved in more than 50% of the sequences are highlighted in blue and red, respectively. In Cg-Def and Cg-BigDef alignments, anionic proregions are in dashed boxes. In Cg-BPI alignment, the central proline-rich region is in dashed boxes.