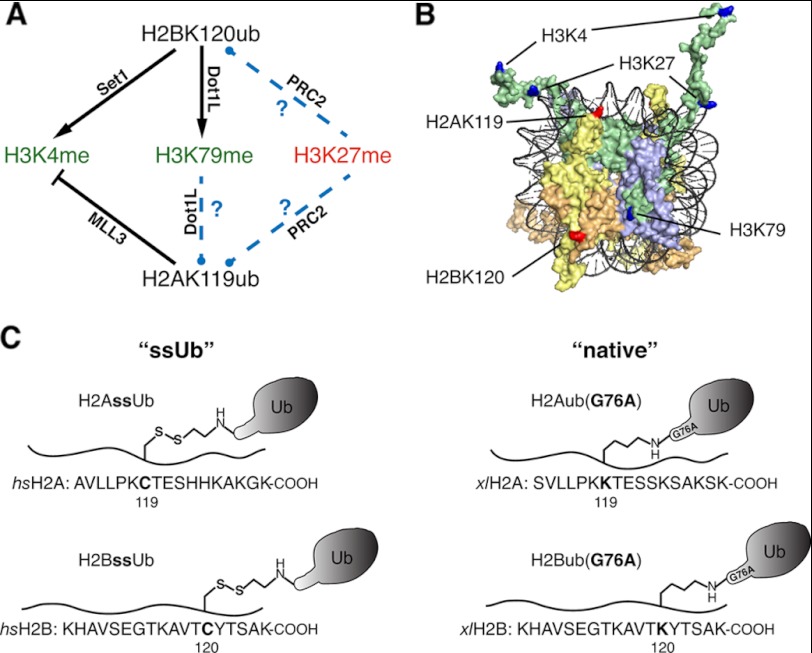
FIGURE 1.
Enzymatic cross-talk between histone ubiquitylation and methylation using designer monoubiquitylated histones. A, the black lines drawn between histone monoubiquitylation and histone methylation sites indicate validated direct enzymatic cross-talk between the indicated PTMs, with the relevant enzyme indicated. Standard and blunt-headed arrows signify enzymatic stimulation and inhibition, respectively, by the modification at the origin of the arrow. Novel cross-talk relationships tested in this study are indicated by dashed blue lines. Methylation states strongly associated with active or repressed transcription are colored green and red, respectively. B, surface rendition of mononucleosome structure (Protein Data Bank code 1KX5). DNA is colored black, and histones H3, H4, H2A, and H2B are colored green, lavender, yellow, and orange, respectively. Lysines site-specifically modified in this study by monoubiquitylation are indicated in red. Lysines colored blue are methylated and are targets for direct enzymatic cross-talk relationships with ubiquitylated histones. C, the four monoubiquitylated histones generated for this study are represented in schematic form. Amino acid sequences of H2A and H2B C termini are shown below a wavy line representing the peptide backbone. Left, ssUb refers to Homo sapiens (hs) H2AssUb and H2BssUb generated by disulfide-mediated ubiquitylation at position 119 in H2A and position 120 in H2B. Right, native refers to Xenopus laevis (xl) H2Aub(G76A) and H2Bub(G76A) generated by semisynthetic expressed protein ligation.