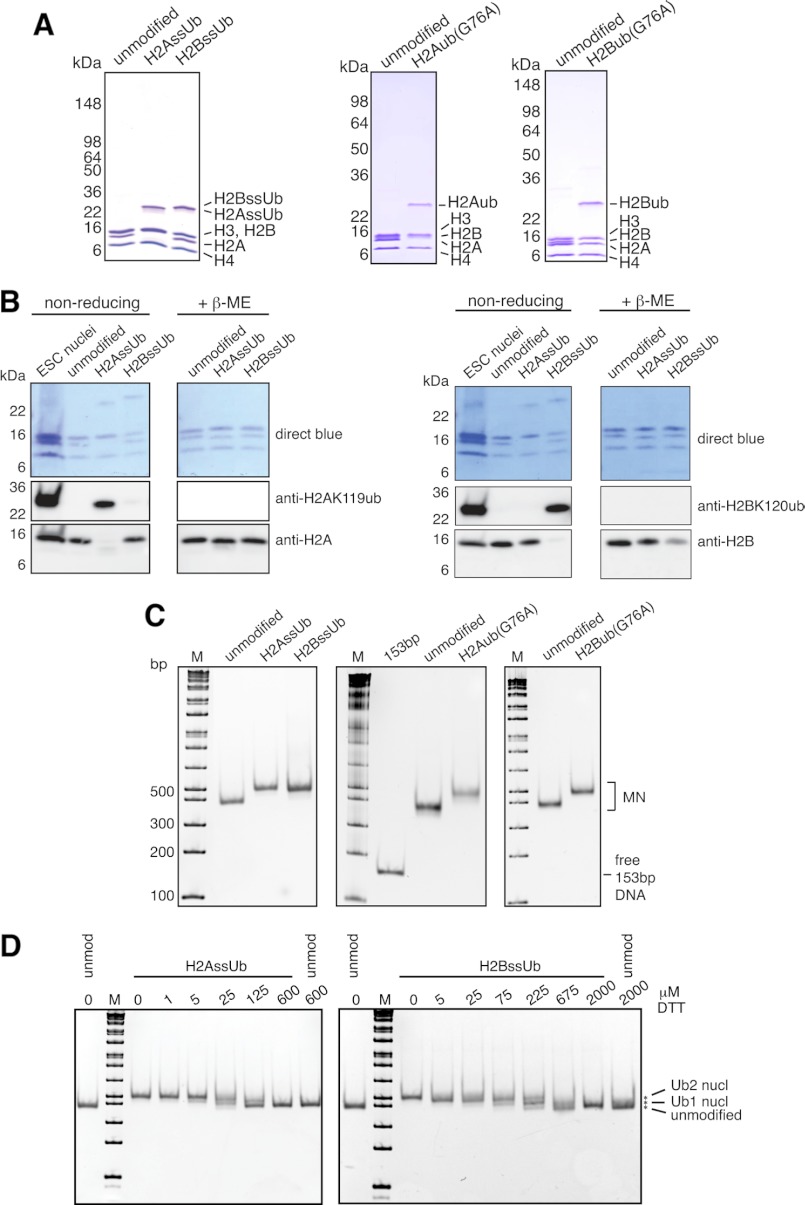
FIGURE 2.
Characterization of designer octamers and mononucleosomes. A and B, octamers assembled from unmodified and monoubiquitylated histones were separated by SDS-PAGE and characterized by Coomassie Blue staining (A) or Western blotting with the indicated antibodies (B). Nuclei from mouse embryonic stem cells (ESC) were used to mark the migration positions of endogenous H2AK119ub and H2BK120ub. β-ME, β-mercaptoethanol. C, human unmodified, H2AssUb, H2BssUb, H2Aub(G76A), and H2Bub(G76A) octamers were assembled into mononucleosomes with 153-bp (601) positioning sequence DNA and separated by native 5% PAGE. Gels were stained with ethidium bromide to confirm the absence of free 153-bp DNA and to assess the structural homogeneity of the assembled mononucleosome (MN) pools. The marker (M) is the 1-kb Plus ladder from New England Biolabs. D, unmodified (unmod nucl), H2AssUb, and H2BssUb mononucleosomes were adjusted to the indicated DTT concentrations and incubated at 37 °C for 3 h before separation by native 5% PAGE and stained with ethidium bromide to assess the stability of the disulfide bond between histone and ubiquitin.