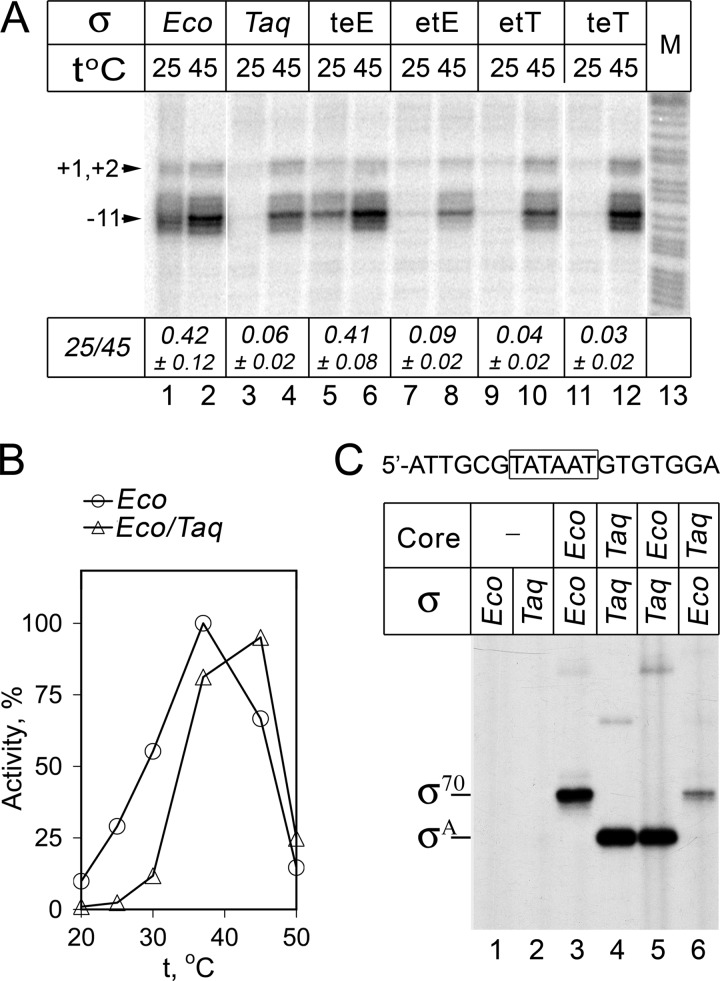
FIGURE 5.
Promoter recognition and opening by RNAP holoenzymes containing Eco core and various σ subunits. A, KMnO4 probing of the lacUV5 promoter complexes. Footprinting was performed at either 25 or 45 °C on a linear DNA fragment containing the lacUV5 promoter; positions of modified thymines in the template promoter strand relative to the starting point of transcription are shown. Lane 13 contains an A+G cleavage marker. The ratio of modification efficiencies at 25 and 45 °C (measured for position −11) for each RNAP is indicated below the figure (averages and standard deviations from two-three independent experiments). B, activities of wild-type Eco and hybrid Eco/Taq RNAPs measured in the reaction of abortive synthesis on the lacUV5 promoter DNA fragment at different temperatures (in percent of the maximal Eco RNAP activity). C, cross-linking of the nontemplate promoter oligonucleotide (shown on the top, the −10 element is boxed) with Eco, Taq, and hybrid Eco/Taq RNAP holoenzymes. The experiment was performed at 25 °C. The positions of cross-linked complexes containing the σ70 and σA subunits are indicated on the left.