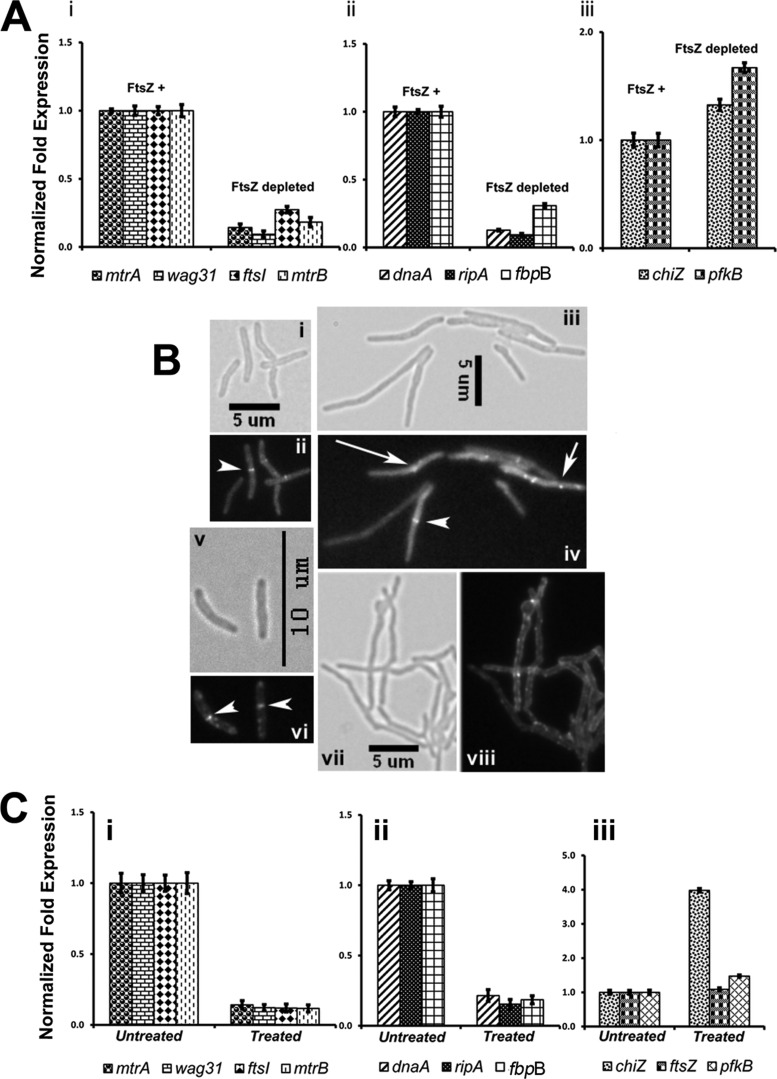
FIGURE 10.
Evaluation of select mtrA targets under conditions affecting MtrB-septal assembly. A, qRT-PCR expression levels of select mtrA targets under conditions affecting FtsZ-septal assembly. Actively growing M. smegmatis FZ-3, ftsZ conditional expression strain, with acetamide (referred to as FtsZ+) was harvested, washed with acetamide-free medium, and grown without acetamide (FtsZ-depleted) or with acetamide (FtsZ+) for 9 h. Total RNA from FtsZ+ and FtsZ-depleted cultures was prepared, and qRT-PCR was performed as described under Fig. 9. Panel i shows expression profiles of mtrA, mtrB, wag31, and ftsI; panel ii shows data for dnaA, ripA, and fbpB; panel iii shows data for chiZ and pfkB. Note: expression levels of chiZ and pfkB in panel iii were modestly affected. B, shown is the effect of mitomycin C exposure on MtrB-GFP and FtsZ-GFP structures. M. smegmatis expressing ftsZ-gfp (panels i–iv) or mtrB-gfp (panels (v–viii) were exposed to 0.5 μg/ml mitomycin C (panels iii, iv, vii, and viii) or grown untreated (panels i, ii, v, and vi) for 6 h, and cells were visualized by microscopy. Panels i, iii, v, and vii show brightfield images, whereas panels ii, iv, vi, and viii show fluorescent images. Septal localizations of FtsZ-GFP (panel ii and iv) and MtrB-GFP (panel vi) are marked with arrowheads. Note that majority of mitomycin C-treated cultures did not contain predominant FtsZ-GFP (panel iv) or MtrB-GFP (panel viii) septal localizations. Aberrant FtsZ-GFP localizations are marked with arrows (panel iv). C, qRT-PCR expression profiles of select MtrA-targets upon exposure to mitomycin C are shown. Total RNA from untreated and 6-h mitomycin C-treated cultures of M. smegmatis was prepared, and qRT-PCR analysis of select targets was performed as described above. Panel i shows the expression profiles of mtrA, wag31, ftsI, and mtrB; panel ii shows data for dnaA, ripA, and fbpB, whereas panel iii shows data for chiZ, ftsZ, and pfkB.