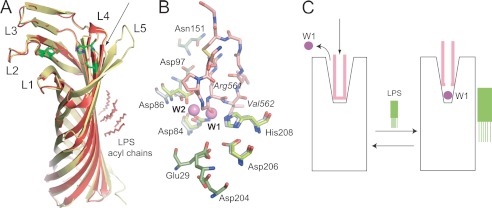
FIGURE 3.
Structure-based model for Pla activation by LPS. A, structural comparison of LPS-free apo-Pla (yellow; PDB ID 2X4M) and LPS-bound apo-Pla (red; PDB ID 2X55). The active site residues Asp84, Asp86, Asp206, and His208 are shown as green stick models for both structures. The conformational changes in the tip of loop L4 that arise from LPS binding are indicated with an arrow. B, close-up of the active site groove of LPS-bound apo-Pla and of the Pla-Alp11 complex. The peptide is colored salmon, with oxygens red and nitrogens blue. The active site residues are shown as sticks in gray (apo-Pla) and in green (Pla-Alp11). The nucleophilic water molecule (W1) and the water molecule W2 between Asp84 and Asp86 that are present in apo-Pla but absent in Pla-Alp11 are shown as purple spheres. Residues within the two acidic binding pockets are shown in green for Pla-Alp11 only. C, schematic model for Pla activation by LPS. In the absence of LPS (left), the activation loop of the substrate (pink) binds deep into the active site groove, displacing the nucleophilic water molecule and rendering the enzyme inactive. Upon binding of LPS, the active site groove narrows (right). When substrate binds, the catalytic water molecule remains in place to cleave the substrate.