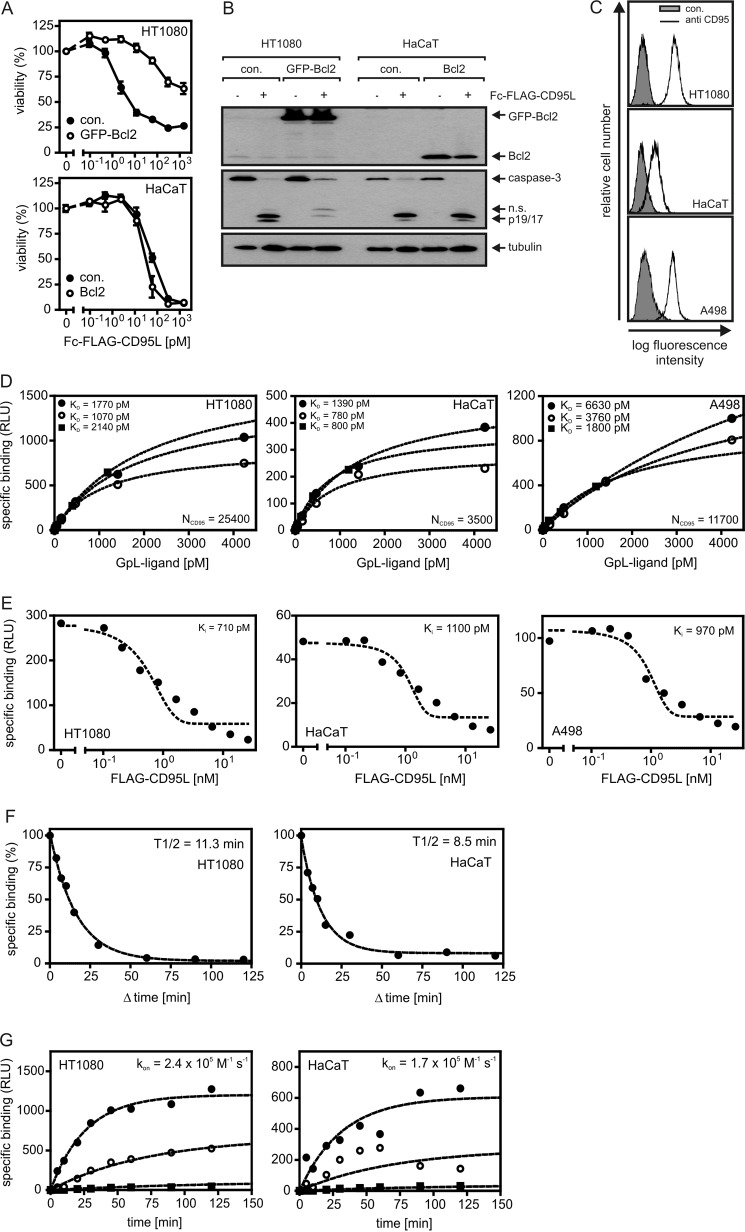
FIGURE 4.
Cellular binding studies with GpL-tagged CD95L variants. A, HT1080 and HaCaT cells and the corresponding transfectants stably expressing Bcl2-GFP or Bcl2 were seeded in 96-well plates (2 × 104 per well) and cultured overnight. The next day cells were stimulated with increasing concentrations of Fc-FLAG-CD95L. After 18 h cell viability was determined via crystal violet staining. B, HT1080 and HaCaT cells and their corresponding Bcl2 transfectants were treated with 200 ng/ml Fc-FLAG-CD95L or remained untreated. After 3 h cells were harvested, and Triton X-100 lysates were separated by SDS-PAGE and analyzed by Western blotting for the indicated proteins. n.s., not significant. C, FACS analysis of cell surface expression of CD95 is shown. D, equilibrium binding studies with GpL-FLAG-CD95L (filled circles), anti-FLAG oligomerized GpL-FLAG-CD95L (open circles), and GpL-Fc-FLAG-CD95L (filled squares) were performed for the indicated cell lines as described under “Experimental Procedures.” CD95 numbers per cell (NCD95) calculated from the experiments shown are indicated. E, for homologous competition experiments, cells were incubated in 24-well plates for 2 h at 37 °C with 200 pm (∼25 ng/ml) GpL-FLAG-CD95L and the indicated concentrations of conventional FLAG-CD95L. Unbound ligand was then removed by 10 washes with ice-cold PBS, and remaining cell associated luciferase activity was determined. F, cells were cultivated in a 24-well plate (2 × 105 cells/well). Half of the wells were pretreated with FLAG-CD95L (0.5 h, 2 μg/ml) to block CD95-pecific binding. Cells of all wells were then incubated for 1 h with 25 ng/ml (∼ 200 pm) of GpL-FLAG-CD95L. To determine dissociation as a function of time, 2 μg/ml FLAG-CD95L were added for the indicated time intervals to blocked and non-blocked samples. Cell associated luciferase activity was determined, and remaining specific binding was calculated for all time points as the difference of blocked and non-blocked samples. G, untreated and FLAG-CD95L-blocked (0.5 h, 1.5 μg/ml) cells were incubated with GpL-FLAG-CD95L (2100 (filled circles), 420 (open circles), and 40 pm (filled squares)) for the indicated times, and specific binding was calculated again as the difference of total (non-blocked samples) and unspecific (blocked samples) binding. The GraphPad Prism5 software was used to fit the association kinetics to obtain the association rate constant. RLU, relative light units.