Background: Zinc-finger antiviral protein (ZAP) is a type I interferon-inducible host antiviral factor against certain viruses, including HIV-1 and Ebola virus.
Results: Glycogen synthase kinase 3β (GSK3β) phosphorylates ZAP, and inhibition of GSK3β reduces ZAP activity.
Conclusion: GSK3β phosphorylation of ZAP modulates its antiviral activity.
Significance: These findings help to better understand how the immune response is regulated.
Keywords: Antiviral Agents, Cellular Regulation, Glycogen Synthase Kinase 3, HIV, Protein Phosphorylation, RNA-binding Protein, RNA Turnover, ZAP, Translational Repression
Abstract
Zinc-finger antiviral protein (ZAP) is a host factor that specifically inhibits the replication of certain viruses, including HIV-1, Ebola virus, and Sindbis virus. ZAP binds directly to specific viral mRNAs and recruits cellular mRNA degradation machinery to degrade the target RNA. ZAP has also been suggested to repress translation of the target mRNA. In this study, we report that ZAP is phosphorylated by glycogen synthase kinase 3β (GSK3β). GSK3β sequentially phosphorylated Ser-270, Ser-266, Ser-262, and Ser-257 of rat ZAP. Inhibition of GSK3β by inhibitor SB216763 or down-regulation of GSK3β by RNAi reduced the antiviral activity of ZAP. These results indicate that phosphorylation of ZAP by GSK3β modulates ZAP activity.
Introduction
The zinc-finger antiviral protein (ZAP)3 was originally isolated as a host factor that inhibits the replication of Moloney murine leukemia virus (MMLV) (1). In addition to MMLV, ZAP inhibits the replication of HIV-1 (2), Ebola virus and Marburg virus (3), and certain alphaviruses such as Sindbis virus, Ross River virus, and Semliki Forest virus (4, 5). ZAP is not a universal antiviral factor because it does not inhibit the replication of viruses such as poliovirus and yellow fever virus (5). Type I interferon treatment or viral infection up-regulates ZAP expression (4, 6, 7), and down-regulation of endogenous ZAP enhances the expression of HIV-1 (2), suggesting that ZAP is an antiviral effector in vivo.
Studies investigating the step at which ZAP inhibits the replication of MMLV have shown that the formation and nuclear entry of viral DNA are not affected. However, the level of viral mRNA in the cytoplasm is dramatically reduced. Further studies have shown that ZAP binds directly to target viral mRNA (2), recruits the cellular polyA ribonuclease (PARN) to remove the poly(A) tail, and recruits the 3′–5′ exoribonuclease complex exosome to degrade the RNA body from the 3′-end (2). DEAD box RNA helicase p72 is required for the optimal antiviral activity of ZAP (8). ZAP also recruits the decapping complex Dcp1-Dcp2 through p72 to remove the cap structure of the target viral mRNA to initiate the degradation of viral mRNA from the 5′-end (2). In addition, it has been suggested that ZAP represses the translation of the target mRNA (2). The mechanisms by which ZAP represses target mRNA translation are not clear yet.
Full-length rat ZAP is composed of 776 amino acids (1) and contains four CCCH-type zinc-finger motifs in the N-terminal domain. The 254-amino acid N-terminal domain of ZAP displays considerable antiviral activity when fused with the Zeocin resistance gene product (NZAP-Zeo) (1). The function of the C-terminal domain of ZAP remains largely elusive.
Glycogen synthase kinase 3β (GSK3β) was initially identified in rabbit skeletal muscle as a serine/threonine kinase that phosphorylates glycogen synthase (9). GSK3 was later shown to have two closely related isoforms (10), GSK3α and GSK3β. For GSK3β to phosphorylate a substrate, a serine or threonine residue of the substrate usually needs to be phosphorylated beforehand. This residue, termed the priming site, is located four or five amino acids C-terminal to the first GSK3β target residue (11, 12). GSK3β sequentially phosphorylates serine/threonine residues from the C terminus to the N terminus. In this study, we demonstrate that GSK3β phosphorylates rat ZAP and modulates its antiviral activity.
EXPERIMENTAL PROCEDURES
Plasmids and Constructs
pcDNA4TO/Myc-ZAP, which expresses Myc-tagged full-length rat ZAP, has been described previously (13). pcDNA4TO/Myc-NZAP332 and pcDNA4TO/Myc-NZAP295 express Myc-tagged NZAP332 (N-terminal amino acids 1–332) and NZAP295 (amino acids 1–295), respectively. The plasmids were generated by replacing the BamHI-NotI fragment of pcDNA4TO/Myc-ZAP with PCR fragments generated using forward primer NZAP-FP (5′-ATCGGATCCGCCACCATGGCAGATCCCGGGGTATG-3′) and reverse primers NZAP332-RP (5′-CTCGAGCGGCCGCCATTCC-3′) and NZAP295-RP (5′-TCGAGCGGCCGCCCTTGAACTTCTGGGT-3′). To generate NZAP295 mutants, pcDNA4TO/Myc-NZAP295 was used as a template for site-directed mutagenesis by overlapping PCR as described previously (14). pGEX-NZAP295 expresses NZAP295 fused with GST at the N terminus (GST-NZAP295). The coding sequence of NZAP295 was PCR-amplified using primers NZAP-FP and NZAP295-RP and cloned into pGEX-5X-3 (GE Healthcare). pGEX-NZAP295(S1–8A), which expresses GST-NZAP295(S1–8A) (where S1–8A represents the substitution of eight serine residues with alanines), was constructed in a similar manner. pMMLV-luc and pRL-TK have been described previously (15).
GSK3β was amplified from a human embryonic kidney cDNA library using forward primer GSK3β-FP (5′-TGAGAATTCTCAGGGCGGCCCAGAACCACCT-3′) bearing an EcoRI site and reverse primer GSK3β-RP (5′-CGGCTCGAGTCAGGTGGAGTTGGAAGCTGAT-3′) bearing an XhoI site and cloned into expression vector pCMV-HA-FLAG (13) using these two sites. To generate a GSK3β-expressing construct that cannot be down-regulated by Gi-5 shRNA, silent mutations were introduced into pCMV-HA-FLAG-GSK3β by overlapping PCR (8). The sequences of the primers used were as follows: GSK3βi5M-FP, 5′-AAGAACCGTGAGTTGCAAATTATGAGAAAG-3′; and GSK3βi5M-RP, 5′-CTTTCTCATAATTTGCAACTCACGGTTCTT-3′ (with the modified nucleotides underlined).
To generate plasmids expressing shRNAs directed against GSK3β, oligonucleotides were synthesized, annealed, and cloned into pSUPER plasmids (Oligoengine) as described previously (8). The sequences of the oligonucleotides used were as follows: GSK3βi3-FP, 5′-GATCCCCACCGCAGAACCTCTTGTTGTTCAAGAGACAACAAGAGGTTCTGCGGTTTTTTA-3′; GSK3βi3-RP, 5′-AGCTTAAAAAACCGCAGAACCTCTTGTTGTCTCTTGAACAACAAGAGGTTCTGCGGTGGG-3′; GSK3βi5-FP, 5′-GATCCCCGAATCGAGAGCTCCAGATCTTCAAGAGAGATCTGGAGCTCTCGATTCTTTTTA-3′; and GSK3βi5-RP, 5′-AGCTTAAAAAGAATCGAGAGCTCCAGATCTCTCTTGAAGATCTGGAGCTCTCGATTCGGG-3′.
Cell Culture
All cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen). 293TRex and 293TRex-ZAP cell lines have been described previously (15). Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Infection and Luciferase Assays
Lentiviral vectors NL4–3-luc and HR′-luc have been described previously (2). To analyze the effect of SB216763 treatment on the antiviral activity of ZAP, 293TRex-ZAP cells were infected with NL4–3-luc or HR′-luc. At 5 h post-infection, tetracycline was added to a final concentration of 1 μg/ml to induce ZAP expression, and SB216763 was added to a final concentration of 10 μm to inhibit GSK3b. At 53 h post-infection, the cells were lysed, and luciferase activity was measured. Fold inhibition was calculated as luciferase activity in cells without tetracycline treatment divided by that in cells treated with tetracycline.
Antibodies
Anti-Myc (Santa Cruz Biotechnology), anti-FLAG (Sigma), and anti-β-actin (Sigma) monoclonal antibodies were obtained commercially. To detect the phosphorylation of ZAP Ser-274 (referred to as S5 in this study), a peptide (CGGPPSPD) in which the serine residue was phosphorylated was synthesized (SciLight Biotechnology), conjugated to keyhole limpet hemocyanin, and used to immunize rabbits to generate a polyclonal antibody. The antibody was affinity-purified with the peptide conjugated to BSA.
In Vitro Protein Kinase Assay
GST-NZAP295 or GST-NZAP295(S1–8A) was expressed in Escherichia coli BL21 and partially purified with glutathione-Sepharose 4B resin (GE Healthcare) following the manufacturer's instructions. The protein was incubated with recombinant GSK3β, CDK1, or CK2 (New England Biolabs) in the presence of 5 μm unlabeled ATP and 10 μCi of [γ-32P]ATP at 30 °C. Reactions were analyzed by SDS-PAGE, followed by autoradiography.
Real-time PCR
293TRex-ZAP cells were infected with NL4–3-luc. At 5 h post-infection, tetracycline was added to induce ZAP expression, and SB216763 was added to inhibit GSK3β. At 53 h post-infection, cells were collected. Ten percent of the cells were lysed to measure luciferase activity. The rest of the cells were used to extract cytoplasmic mRNA, followed by reverse transcription. nef-luc mRNA levels were measured by TaqMan real-time PCR as described previously (2).
RESULTS
ZAP Is Phosphorylated
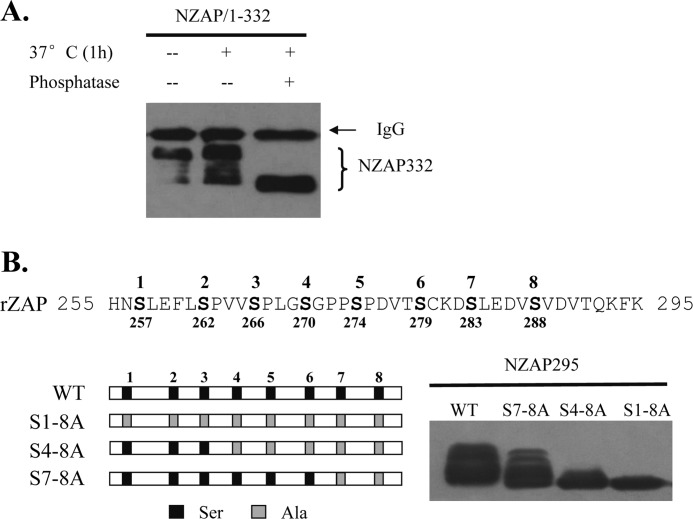
When the 332-amino acid N-terminal domain of ZAP (NZAP332) expressed in HEK293 cells was analyzed by Western blotting, multiple bands were detected (Fig. 1A). We speculated that their appearance might result from phosphorylation of the protein. To test this idea, NZAP332 was immunoprecipitated and treated with alkaline phosphatase, followed by Western blot analysis. Alkaline phosphatase treatment abolished the mobility shift of NZAP332 (Fig. 1A), indicating that NZAP332 is phosphorylated and that the phosphorylation leads to a mobility shift.
FIGURE 1.
ZAP is phosphorylated. A, treatment of ZAP with alkaline phosphatase changes the mobility of ZAP. A Myc-NZAP332-expressing vector was transiently expressed in HEK293 cells. The protein was immunoprecipitated with protein G resin and anti-Myc antibody and treated with alkaline phosphatase or mock-treated. Samples were analyzed by Western blotting. B, substitutions of the serines between amino acids 255 and 295 with alanines alter the mobility of ZAP. The sequence of amino acids 255–295 of rat ZAP (rZAP) is shown (upper), and the substitutions of serines with alanines are schematically presented (lower left). The Myc-tagged NZAP295 proteins indicated were transiently expressed in HEK293 cells and analyzed by Western blotting (lower right). The numbers above the sequence are the numbers used to identify the serines studied in this work.
There are eight serine residues in ZAP in the region of amino acids 255–295 (numbered 1–8 from the N terminus in this report (Fig. 1B)), which are possible phosphorylation sites. There are also some serine residues in the region of amino acids 296–332. We focused on the serine residues in the region of amino acids 255–295 and thus constructed NZAP295, which covers amino acids 1–295. Substitutions of the eight serine residues with alanines (S1–8A) abolished the mobility shift (Fig. 1B), whereas substitution of the fourth through eighth serines (S4–8A) removed most but not all of the mobility shift, and mutation of only the seventh and eighth serines (S7–8A) changed the mobility shift only modestly (Fig. 1B). These results imply that at least some of these serine residues are phosphorylated.
GSK3β Phosphorylates ZAP
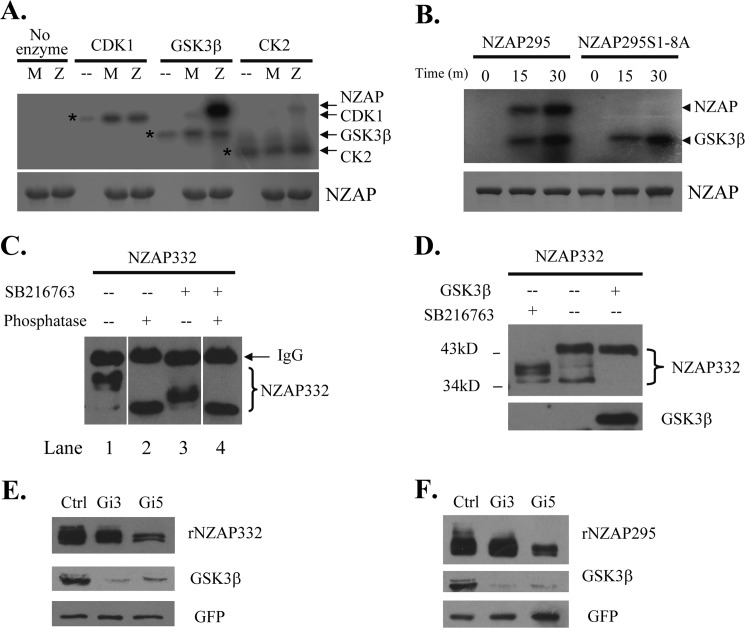
Sequence analysis of amino acids 1–295 of rat ZAP using NetPhosK software (16) predicted that ZAP might be a substrate of GSK3β, as it contains several potential GSK3β phosphorylation motifs, (S/T)XXX(S/T). To test this idea, bacterially expressed GST-NZAP295 or GST-NZAP295(S1–8A) was incubated with recombinant GSK3β and [γ-32P]ATP in vitro. Serine/threonine kinases CDK1 and CK2 were used as specificity controls. Indeed, GSK3β phosphorylated the wild-type protein but not the GST-NZAP295(S1–8A) mutant (Fig. 2A). In contrast, CDK1 failed to phosphorylate GST-NZAP295, and phosphorylation of the protein by CK2 was very modest (Fig. 2A). Detection of comparable autophosphorylation of the kinases indicates that comparable amounts of active enzymes existed in the reactions (Fig. 2A). Phosphorylation of GST-NZAP295 by GSK3β increased with prolonged reaction times (Fig. 2B). In contrast, phosphorylation of GST-NZAP295(S1–8A) was not detected (Fig. 2B). These results suggest that GSK3β phosphorylates at least some of the eight serine residues in vitro.
FIGURE 2.
ZAP is phosphorylated by GSK3β. A and B, recombinant NZAP295 is phosphorylated by GSK3β in vitro. Recombinant proteins GST-NZAP295 and GST-NZAP295(S1–8A) were bacterially expressed, partially purified, and incubated with GSK3β, CDK1, or CK2 in the presence of [γ-32P]ATP at 30 °C. The reactions were analyzed by SDS-PAGE and autoradiography (upper panel). That equal amounts of GST-NZAP295 and GST-NZAP295(S1–8A) were used was confirmed by Coomassie Blue staining (lower panel). A, GST-NZAP295 and GST-NZAP295(S1–8A) were incubated with the enzymes indicated for 30 min. The positions of the kinases are indicated by asterisks. −, no recombinant ZAP proteins were added to the reaction; M, GST-NZAP295(S1–8A); Z, GST-NZAP295. B, GST-NZAP295 and GST-NZAP295(S1–8A) were incubated with GSK3β for the lengths of time indicated. C, inhibition of GSK3β reduces ZAP phosphorylation. The plasmid expressing Myc-tagged NZAP332 was transfected into HEK293 cells. At 6 h post-transfection, cells were treated with SB216763 or mock-treated. The proteins were immunoprecipitated, incubated with alkaline phosphatase at 37 °C for 30 min, and analyzed by Western blotting. D, the plasmid expressing Myc-tagged NZAP332 was cotransfected with the plasmid expressing GSK3β. At 6 h post-transfection, the cells were mock-treated (−) or treated with SB216763 (+). Cells were lysed at 48 h post-transfection, and the lysates were analyzed by Western blotting with anti-Myc antibody to detect NZAP332 or with anti-FLAG antibody to detect GSK3β. E and F, a plasmid expressing the shRNA directed against GSK3β (Gi-3 or Gi-5) was transfected into HEK293 cells. At 24 h post-transfection, puromycin was added to a final concentration of 3 μg/ml to select for transfected cells. Three days later, a second transfection was done under the same conditions. At 24 h after the second transfection, a plasmid expressing Myc-tagged recombinant NZAP332 (rNZAP332) (E) or recombinant NZAP295 (rNZAP295) (F) was transfected. A plasmid expressing Myc-tagged GFP was included as a control for transfection efficiency and sample handling. At 24 h post-transfection, the cells were lysed, and the proteins were analyzed by Western blotting. Ctrl, control.
To test whether GSK3β phosphorylates ZAP in vivo, we treated cells with SB216763, a small-molecule compound that specifically inhibits the kinase activity of GSK3β (17). As expected, treatment of cells with SB216763 resulted in a dramatic reduction of the phosphorylation of Myc-tagged NZAP332 (Fig. 2C, compare lanes 1 and 3). Notably, when the lysate of SB216763-treated cells was incubated with alkaline phosphatase, NZAP332 phosphorylation was further reduced (Fig. 2C, compare lanes 3 and 4), suggesting that NZAP332 is also phosphorylated by other kinase(s). In addition, when GSK3β was overexpressed, bands of less phosphorylated NZAP332 were absent (Fig. 2D). To further demonstrate that ZAP is phosphorylated by GSK3β, shRNAs directed against GSK3β (Gi-3 and Gi-5) were constructed. Consistently, down-regulation of endogenous GSK3β by the shRNAs led to reduction of NZAP295 and NZAP332 phosphorylation (Fig. 2, E and F). Taken together, these results strongly indicate that GSK3β phosphorylates ZAP in vivo.
Mapping Serine Residues of ZAP Phosphorylated by GSK3β
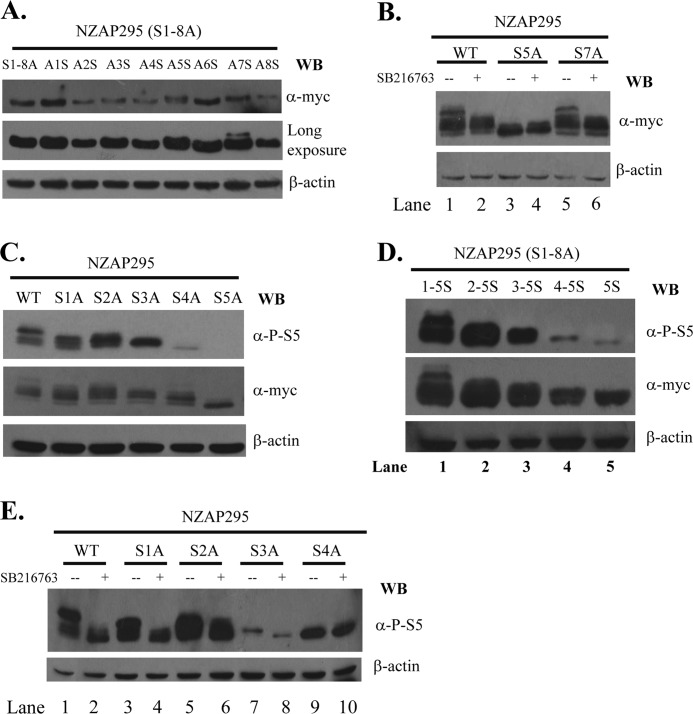
To identify the serine residues of NZAP295 phosphorylated by GSK3β, we individually substituted the alanines of NZAP295(S1–8A) with serines and analyzed the phosphorylation of these mutants by mobility shift assays. Only substitution of the fifth and seventh alanines with serine caused marked mobility shifts (Fig. 3A). When S5 of NZAP295 was replaced with alanine, most of the phosphorylated bands disappeared (Fig. 3B, compare lanes 1 and 3). In comparison, substitution of S7 of NZAP295 with alanine had little effect on the phosphorylation (Fig. 3B, compare lanes 1 and 5). When cells were treated with SB216763, phosphorylation of NZAP295(S7A) was dramatically reduced (Fig. 3B, compare lanes 1 and 2 and lanes 5 and 6). In contrast, SB216763 had no effect on NZAP295(S5A) (Fig. 3B, compare lanes 3 and 4). These results suggest that S5 is critical for GSK3β to phosphorylate ZAP and could be either the priming site or a phosphorylation site of GSK3β.
FIGURE 3.
Mapping serine residues phosphorylated by GSK3β. A, C, and D, the proteins indicated were transiently expressed in HEK293 cells, subjected to SDS-PAGE, and detected by Western blotting (WB) with the antibodies indicated. B and E, a plasmid expressing the proteins indicated was transfected into HEK293 cells. At 6 h post-transfection, the cells were mock-treated (−) or treated with SB216763 (+). Cells were lysed at 48 h post-transfection, and the lysates were analyzed by Western blotting with the antibodies indicated.
To detect phosphorylation of S5, we developed a polyclonal antibody against a peptide containing phosphorylated S5 (see “Experimental Procedures”). The antibody (anti-P-S5) specifically detected ZAP with phosphorylated S5 but not the S5A mutant (Fig. 3C, upper panel). Fewer bands were detected with this antibody than with the anti-Myc antibody, which detects all forms of the protein (Fig. 3C, compare upper and middle panels), suggesting that some of the protein is not phosphorylated at S5. To further map the GSK3β phosphorylation sites of ZAP, we individually substituted the serines of NZAP295 with alanine and analyzed their phosphorylation status. The S5A mutation removed most of the phosphorylation of the protein (Fig. 3C). In comparison, substitution of S1–S4 had less effect, with substitution of the residue at the N terminus having less effect than substitution of the residue at the C terminus (Fig. 3C). This pattern suggests that S5 is the priming site and that GSK3β sequentially phosphorylates S4 to S1. To substantiate this notion, we substituted the alanines of NZAP295(S1–8A) with serine one by one. When the fifth alanine was replaced with serine (NZAP295(S1–8A)S5), a single band was detected by anti-P-S5 antibody (Fig. 3D, upper panel, lane 5). When the fifth and fourth alanines were replaced with serines (NZAP295(S1–8A)S4-S5), the mobility of the protein was slower than that of NZAP295(S1–8A)S5 (Fig. 3D, upper panel, compare lanes 4 and 5), suggesting that both S4 and S5 are phosphorylated. Replacement of the third, second, and first alanines with serine sequentially increased the phosphorylation of the protein (Fig. 3D), indicating that these residues are phosphorylated. To test whether these residues are phosphorylated by GSK3β, cells expressing the Ser-to-Ala single mutants were treated with SB216763. The inhibitor had no effect on the phosphorylation of NZAP295(S4A) (Fig. 3E, compare lanes 9 and 10) but decreased the phosphorylation of NZAP295(S3A) (compare lanes 7 and 8). These results are consistent with the notion that S4 is the first residue phosphorylated by GSK3β and that its phosphorylation is required for the phosphorylation of S3 by GSK3β. Substitution of S2 with alanine reduced phosphorylation (Fig. 3E, compare lanes 1 and 5), and SB216763 treatment also reduced phosphorylation (compare lanes 5 and 6). A similar phenomenon was also observed with S1 (Fig. 3E, compare lanes 1 and 3 and lanes 3 and 4). Taken together, these results show that GSK3β sequentially phosphorylates S4 to S1, with S5 as the priming site.
Phosphorylation by GSK3β Enhances Antiviral Activity of ZAP
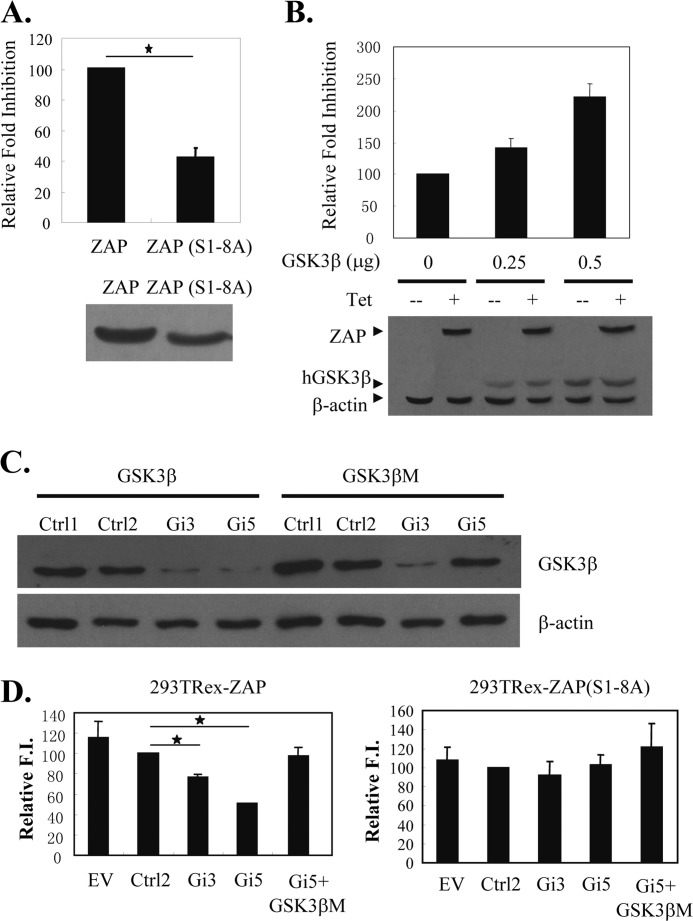
To investigate the effect of GSK3β phosphorylation on ZAP function, we first compared the antiviral activity of ZAP and ZAP(S1–8A) against MMLV-luc (15). Compared with wild-type ZAP, the mutant displayed reduced antiviral activity (Fig. 4A), suggesting that phosphorylation is required for optimal activity of ZAP. Furthermore, overexpression of GSK3β in 293TRex-ZAP cells, which express ZAP upon tetracycline treatment, increased ZAP activity in a dose-dependent manner (Fig. 4B).
FIGURE 4.
Overexpression of GSK3β enhances ZAP activity, whereas down-regulation of GSK3β reduces ZAP activity. A, 293TRex cells were cotransfected with a plasmid expressing wild-type ZAP or ZAP(S1–8A) and the ZAP-responsive reporter pMMLV-luc. At 6 h post-transfection, cells were mock-treated or treated with 1 μg/ml tetracycline to induce ZAP and ZAP(S1–8A) expression. Cells were lysed, and luciferase activities were measured at 48 h post-transfection. Relative -fold inhibition was calculated as the firefly luciferase activity with ZAP divided by that with ZAP(S1–8A). Data are presented as means ± S.D. of three independent experiments (upper panel). The expression of ZAP and ZAP(S1–8A) was confirmed by Western blotting (lower panel). B, 293TRex-ZAP cells were cotransfected with the indicated amounts of the GSK3β-expressing plasmid, the ZAP-responsive reporter pMMLV-luc, and the internal control pRL-TK. At 6 h post-transfection, cells were mock-treated or treated with 1 μg/ml tetracycline to induce ZAP expression. Cells were lysed, and luciferase activities were measured at 48 h post-transfection. The firefly luciferase activity expressed from pMMLV-luc was normalized by the Renilla luciferase activity expressed from pRL-TK. -Fold inhibition was calculated as the normalized luciferase activity in the mock-treated cells divided by that in the tetracycline-treated cells. Relative -fold inhibition was calculated as the -fold inhibition with GSK3β divided by that without GSK3β (upper panel). Data are presented as means ± S.D. of three independent experiments. The expression of ZAP and GSK3β was confirmed by Western blotting (lower panel). C, plasmids expressing the indicated FLAG-tagged proteins and shRNAs were cotransfected into HEK293 cells. Protein expression levels were measured by Western blotting at 48 h post-transfection. D, 293TRex-ZAP cells (left panel) and 293TRex-ZAP(S1–8A) (right panel) were transfected with the plasmids indicated together with pMMLV-luc and pRL-TK. Relative -fold inhibition (F.I.) was calculated as described for B. Data are presented as means ± S.D. of three independent experiments. GSK3βM is the GSK3β-expressing plasmid that is not targeted by Gi-5. hGSK3β, human GSK3β; EV, pSUPER.retro empty vector; Ctrl, control. *, p < 0.05.
To test whether endogenous GSK3β modulates ZAP activity, endogenous GSK3β was down-regulated by RNAi, and the effect on the antiviral activity of ZAP against MMLV-luc was evaluated. To confirm the specificity of the shRNA (Gi-5) to target GSK3β, a GSK3β-expressing plasmid (GSK3βM) that cannot be targeted by Gi-5 was constructed (Fig. 4C). Down-regulation of GSK3β reduced the antiviral activity of ZAP, and the reduction by Gi-5 was rescued by coexpression of GSK3βM (Fig. 4D, left panel). In contrast, down-regulation of GSK3β had little effect of the antiviral activity of ZAP(S1–8A) (Fig. 4D, right panel).
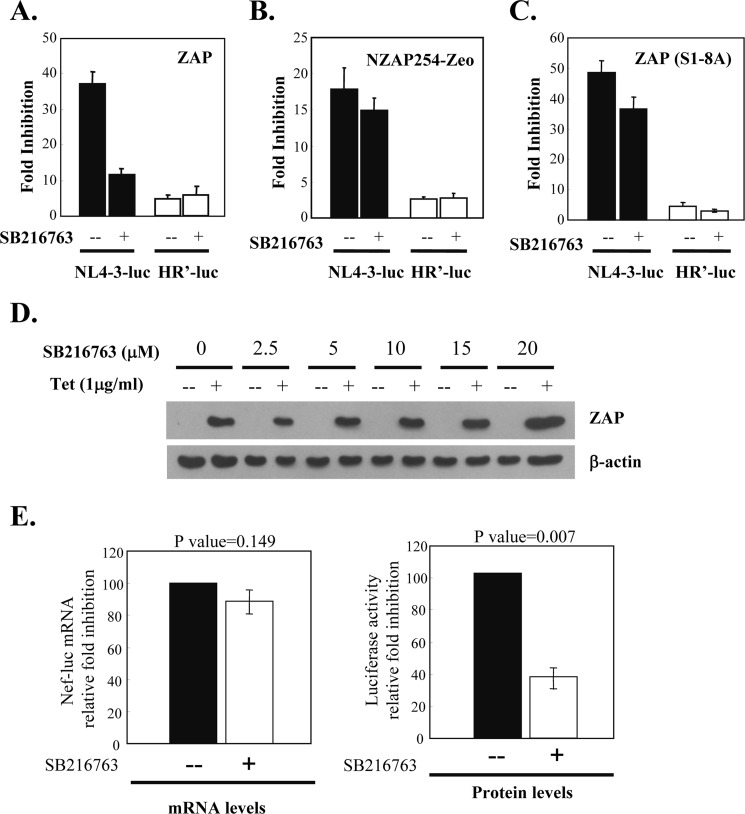
To evaluate the effect of GSK3β-mediated phosphorylation on the antiviral activity of ZAP against HIV-1 pseudovirus, ZAP-expressing cells were treated with SB216763 to inhibit GSK3β. The cells were challenged with the HIV-1 vector NL4–3-luc, which is inhibited by ZAP (2), or with HR′-luc, which is not sensitive to ZAP (2). SB216763 treatment did not affect the expression of ZAP (Fig. 5D) but dramatically reduced the activity of ZAP against NL4–3-luc (Fig. 5A). In contrast, SB216763 treatment did not affect the expression of HR′-luc (Fig. 5A). Furthermore, SB216763 treatment had little effect on the antiviral activity of NZAP-Zeo (Fig. 5B) and ZAP(S1–8A) (Fig. 5C), which do not contain the GSK3β phosphorylation sites. It is worth mentioning that the -fold inhibition of ZAP(S1–8A) against NL4–3-luc was higher than that of wild-type ZAP. Because the antiviral activity of ZAP is dependent on its expression level (3), and fold inhibition is calculated as luciferase activity in the mock-treated cells divided by that in the tetracycline-treated cells, fold inhibition not only reflects the intrinsic antiviral activity but also the relative expression levels of ZAP. The higher fold inhibition of ZAP(S1–8A) could be just a clonal effect. SB216763 treatment of 293TRex-ZAP cells did not affect the expression of ZAP (Fig. 5D). Hence, the effect of SB216763 on the fold inhibition of a particular protein should just reflect its effect on the intrinsic antiviral activity.
FIGURE 5.
Inhibition of GSK3β reduces ZAP antiviral activity. A–C, 293TRex-ZAP, 293TRex-NZAP-Zeo, and 293TRex-ZAP(S1–8A) cells, respectively, were infected with NL4–3-luc or HR′-luc. At 5 h post-infection, cells were treated with tetracycline to induce ZAP expression and with SB216763 to inhibit GSK3β. At 53 h post-infection, cells were lysed, and luciferase activity was measured. -Fold inhibition was calculated as luciferase activity in the untreated cells divided by luciferase activity in the tetracycline-treated cells. Data are presented as means ± S.D. of three independent experiments. D, SB216763 treatment does not affect the expression of ZAP. 293TRex-ZAP cells were treated with the amounts of SB216763 indicated and then mock-treated (−) or treated with tetracycline (Tet; +) to induce ZAP expression. ZAP expression levels were analyzed by Western blotting. E, 293TRex-ZAP cells were infected with NL4–3-luc. At 5 h post-infection, cells were treated with tetracycline to induce ZAP expression and with SB216763 to inhibit GSK3β. At 53 h post-infection, 10% of the cells were lysed to measure luciferase activity, and the rest of the cells were used to extract cytoplasmic RNA. The RNA was reverse-transcribed, followed by measurement of the levels of the reporter nef-luc and gapdh mRNAs by real-time PCR. RNA -fold inhibition was calculated as the nef-luc mRNA level normalized by the gapdh mRNA level in the mock-treated cells divided by that in the tetracycline-treated cells (left panel). Protein -fold inhibition was calculated as the luciferase activity in the mock-treated cells divided by that in the tetracycline-treated cells (right panel). Data are presented as means ± S.D. of three independent experiments.
It has been suggested that ZAP can both repress the translation and promote the degradation of its target mRNA (2). To test which aspects of the antiviral activity of ZAP are affected by GSK3β phosphorylation, we analyzed whether treatment of 293TRex-ZAP cells with SB216763 affects ZAP-mediated reduction in the reporter mRNA level. The data show that SB216763 treatment reduced ZAP activity to inhibit reporter protein expression but had little effect on ZAP activity to promote reporter mRNA degradation (Fig. 5E). These results suggest that phosphorylation of ZAP by GSK3β affects its ability to repress the translation of target mRNA but not its ability to promote target mRNA degradation. Collectively, these results indicate that phosphorylation by GSK3β is required for the optimal functioning of ZAP.
DISCUSSION
ZAP inhibits the replication of certain viruses by binding to specific viral mRNAs and recruiting cellular RNA degradation machinery to degrade target RNA. In addition, it has been suggested that ZAP represses the translation of target viral mRNAs (2). Here, we have shown that GSK3β phosphorylates some of the serine residues of ZAP and that phosphorylation by GSK3β enhances the antiviral activity of ZAP. Furthermore, our results suggest that phosphorylation of ZAP by GSK3β affects its ability to repress target mRNA translation without affecting its ability to promote target mRNA degradation (Fig. 5E). This notion is supported by our preliminary results that mutation of S1–S8 or SB216763 treatment of ZAP reduced its interaction with a cellular factor involved in translational repression but did not affect its interaction with components of the mRNA degradation machinery, such as exosome component Rrp46 and PARN.4 Further investigation is needed to detail the mechanisms by which phosphorylation of ZAP by GSK3β modulates its activity.
Ser-270, Ser-266, Ser-262, and Ser-257 (S1–S4 in this work) were mapped as the phosphorylation sites of GSK3β, with Ser-274 (S5) as the priming site. Single mutation of Ser-274 led to dramatic changes in NZAP phosphorylation. It remains to be determined which kinase phosphorylates Ser-274. Bacterially expressed recombinant ZAP, which presumably lacks the phosphorylated priming site, was phosphorylated by GSK3β in in vitro kinase assays. One possible explanation is that GSK3β can execute phosphorylation without phosphorylation of the priming site, but when the priming site is phosphorylated, GSK3β works more efficiently. Similar observations have also been reported for the phosphorylation of tau and β-catenin by GSK3β (18–20).
GSK3β plays regulatory roles in various diseases (21), including diabetes (22, 23), Alzheimer disease (24, 25), bipolar mood disorder (26), and cancer (27). GSK3β is also involved in innate and adaptive immune responses (28–30). Lithium has been used as a GSK3β inhibitor in the treatment of bipolar disorder. Other GSK3β inhibitors are being tested for the treatment of Alzheimer disease (31–33), type 2 diabetes (32, 34), and osteoporosis (31). Our results showing that inhibition of GSK3β compromises the antiviral activity of ZAP suggest that precautions should be taken in the clinical use of GSK3β inhibitors.
This work was supported by Ministry of Science and Technology 973 Program Grant 2012CB910203, National Science Foundation Grants 30530020 and 81028011, and Ministry of Health of China Grant 2012ZX10001-006 (to G. G.).
L. Sun and G. Gao, unpublished data.
- ZAP
- zinc-finger antiviral protein
- MMLV
- Moloney murine leukemia virus
- GSK3β
- glycogen synthase kinase 3β
- luc
- luciferase.
REFERENCES
- 1. Gao G., Guo X., Goff S. P. (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc-finger protein. Science 297, 1703–1706 [DOI] [PubMed] [Google Scholar]
- 2. Zhu Y., Chen G., Lv F., Wang X., Ji X., Xu Y., Sun J., Wu L., Zheng Y. T., Gao G. (2011) Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 15834–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Müller S., Möller P., Bick M. J., Wurr S., Becker S., Günther S., Kümmerer B. M. (2007) Inhibition of filovirus replication by the zinc-finger antiviral protein. J. Virol. 81, 2391–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y., Burke C. W., Ryman K. D., Klimstra W. B. (2007) Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81, 11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bick M. J., Carroll J. W., Gao G., Goff S. P., Rice C. M., MacDonald M. R. (2003) Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77, 11555–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang N., Dong Q., Li J., Jangra R. K., Fan M., Brasier A. R., Lemon S. M., Pfeffer L. M., Li K. (2010) Viral induction of the zinc-finger antiviral protein is IRF3-dependent but NF-κB-independent. J. Biol. Chem. 285, 6080–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacDonald M. R., Machlin E. S., Albin O. R., Levy D. E. (2007) The zinc-finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J. Virol. 81, 13509–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen G., Guo X., Lv F., Xu Y., Gao G. (2008) p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc. Natl. Acad. Sci. U.S.A. 105, 4352–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Embi N., Rylatt D. B., Cohen P. (1980) Glycogen synthase kinase 3 from rabbit skeletal muscle. Separation from cyclic AMP-dependent protein kinase and phosphorylase kinase. Eur. J. Biochem. 107, 519–527 [PubMed] [Google Scholar]
- 10. Woodgett J. R. (1990) Molecular cloning and expression of glycogen synthase kinase 3/factor A. EMBO J. 9, 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fiol C. J., Mahrenholz A. M., Wang Y., Roeske R. W., Roach P. J. (1987) Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262, 14042–14048 [PubMed] [Google Scholar]
- 12. Frame S., Cohen P., Biondi R. M. (2001) A common phosphate-binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 13. Guo X., Ma J., Sun J., Gao G. (2007) The zinc-finger antiviral protein recruits the RNA-processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U.S.A. 104, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X., Lv F., Gao G. (2010) Mutagenesis analysis of the zinc-finger antiviral protein. Retrovirology 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo X., Carroll J. W., Macdonald M. R., Goff S. P., Gao G. (2004) The zinc-finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc-finger motifs. J. Virol. 78, 12781–12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blom N., Gammeltoft S., Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 17. Coghlan M. P., Culbert A. A., Cross D. A., Corcoran S. L., Yates J. W., Pearce N. J., Rausch O. L., Murphy G. J., Carter P. S., Roxbee Cox L., Mills D., Brown M. J., Haigh D., Ward R. W., Smith D. G., Murray K. J., Reith A. D., Holder J. C. (2000) Selective small-molecule inhibitors of glycogen synthase kinase 3 modulate glycogen metabolism and gene transcription. Chem. Biol. 7, 793–803 [DOI] [PubMed] [Google Scholar]
- 18. Ishiguro K., Omori A., Takamatsu M., Sato K., Arioka M., Uchida T., Imahori K. (1992) Phosphorylation sites on tau by tau protein kinase I, a bovine-derived kinase generating an epitope of paired helical filaments. Neurosci. Lett. 148, 202–206 [DOI] [PubMed] [Google Scholar]
- 19. Imahori K., Uchida T. (1997) Physiology and pathology of tau protein kinases in relation to Alzheimer disease. J. Biochem. 121, 179–188 [PubMed] [Google Scholar]
- 20. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 21. Cohen P., Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 22. Eldar-Finkelman H. (2002) Glycogen synthase kinase 3: an emerging therapeutic target. Trends Mol. Med. 8, 126–132 [DOI] [PubMed] [Google Scholar]
- 23. Lochhead P. A., Coghlan M., Rice S. Q., Sutherland C. (2001) Inhibition of GSK3 selectively reduces glucose-6-phosphatase and phosphatase and phosphoenolypyruvate carboxykinase gene expression. Diabetes 50, 937–946 [DOI] [PubMed] [Google Scholar]
- 24. Grimes C. A., Jope R. S. (2001) The multifaceted roles of glycogen synthase kinase 3β in cellular signaling. Prog. Neurobiol. 65, 391–426 [DOI] [PubMed] [Google Scholar]
- 25. Hernández F., Avila J. (2008) The role of glycogen synthase kinase 3 in the early stages of Alzheimer disease. FEBS Lett. 582, 3848–3854 [DOI] [PubMed] [Google Scholar]
- 26. Jope R. S. (1999) Anti-bipolar therapy: mechanism of action of lithium. Mol. Psychiatry 4, 117–128 [DOI] [PubMed] [Google Scholar]
- 27. Manoukian A. S., Woodgett J. R. (2002) Role of glycogen synthase kinase 3 in cancer: regulation by Wnts and other signaling pathways. Adv. Cancer Res. 84, 203–229 [DOI] [PubMed] [Google Scholar]
- 28. Beurel E., Michalek S. M., Jope R. S. (2009) Innate and adaptive immune responses regulated by glycogen synthase kinase 3 (GSK3). Trends Immunol. 31, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin M., Rehani K., Jope R. S., Michalek S. M. (2005) Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 6, 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lei C. Q., Zhong B., Zhang Y., Zhang J., Wang S., Shu H. B. (2010) Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 33, 878–889 [DOI] [PubMed] [Google Scholar]
- 31. Luna-Medina R., Cortes-Canteli M., Sanchez-Galiano S., Morales-Garcia J. A., Martinez A., Santos A., Perez-Castillo A. (2007) NP031112, a thiadiazolidinone compound, prevents inflammation and neurodegeneration under excitotoxic conditions: potential therapeutic role in brain disorders. J. Neurosci. 27, 5766–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Medina M., Castro A. (2008) Glycogen synthase kinase 3 (GSK3) inhibitors reach the clinic. Curr. Opin. Drug Discov. Devel. 11, 533–543 [PubMed] [Google Scholar]
- 33. Hernández F., Nido J. D., Avila J., Villanueva N. (2009) GSK3 inhibitors and disease. Mini-Rev. Med. Chem. 9, 1024–1029 [DOI] [PubMed] [Google Scholar]
- 34. Wagman A. S., Johnson K. W., Bussiere D. E. (2004) Discovery and development of GSK3 inhibitors for the treatment of type 2 diabetes. Curr. Pharm. Des. 10, 1105–1137 [DOI] [PubMed] [Google Scholar]