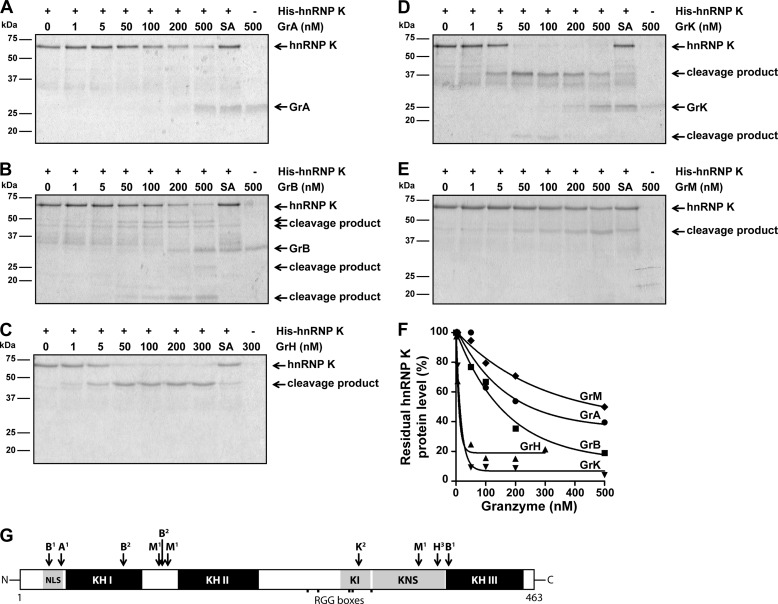
FIGURE 2.
HnRNP K is a direct pan-granzyme substrate. A, purified His-hnRNP K (1 μg, ∼750 nm) was incubated with increasing concentrations of GrA, (B) GrB, (C) GrH, (D) GrK, (E) GrM, or their corresponding catalytically inactive control granzyme (SA) (300 nm for GrH-SA and 500 nm for the other control granzymes) in a buffer containing 50 mm Tris, pH 7.4, and 150 mm NaCl for 16 h at 37 °C, subjected to SDS-PAGE, and all proteins were stained with Simply Blue. F, band intensities were quantified and plotted with His-hnRNP K without granzyme set at 100%. A non-linear regression curve (one phase exponential decay) was fitted, and the concentration of granzyme required to cleave 50% hnRNP K was determined for each granzyme (GrA, 249 nm; GrB, 146 nm; GrH, 9.5 nm; GrK, 13 nm; GrM, 499 nm). G, schematic overview of granzyme cleavage sites within hnRNP K. Cleavage sites were identified by 1) proteomic screens (25–27, 29, 30), 2) site-directed mutagenesis and validated by immunoblotting, or amino acid regions harboring a potential granzyme cleavage site were predicted by 3) immunoblotting using antibodies directed against the mid region as well as the absolute N or C terminus of hnRNP K. (NLS, nuclear localization signal; KH, K homology domain; KI, K interactive region; KNS, K nuclear shuttling signal; RGG, arginine-glycine-glycine)