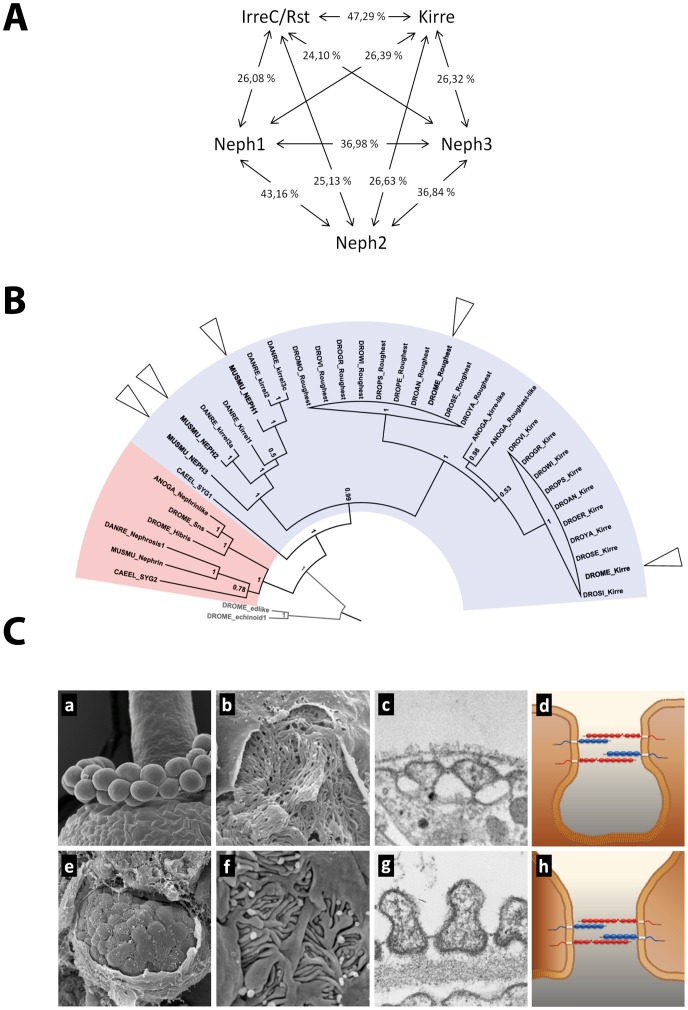
Figure 1. The irre cell recognition module (IRM) is conserved across species.
A. Similarity network of Neph1–3, Kirre and IrreC/Rst generated with T-Coffee multiple sequence alignment algorithm. Values represent full length protein similarity in percent. B. Protein tree of the Neph (blue) and Nephrin (red) protein families as present in Mus musculus, Danio rerio, Caenorhabditis elegans, Anopheles gambiae and several Drosophila species (Drosophila mojavensis (DROMO), Drosophila virilis (DROVI), Drosophila grimshawi (DROGR), Drosophila willistoni (DROWI), Drosophila pseudoobscura (DROPS), Drosophila persimilis (DROPE), Drosophila ananassae (DROAN), Drosophila melanogaster (DROME), Drosophila sechellia (DROSE), Drosophila yakuba (DROYA). Arrowheads mark the proteins investigated in this study. C. Structural comparison of M. musculus kidney glomerulus (e), podocytes (f) and slit diaphragm (g) with GCNs of D. melanogaster (a). The rough surface of the GCN underneath the basement membrane (b) is formed by the nephrocyte diaphragm (c). In contrast to the mammalian slit diaphragm which is formed between neighboring podocytes (h), the Drosophila nephrocyte diaphragm is formed within one GCN (d).