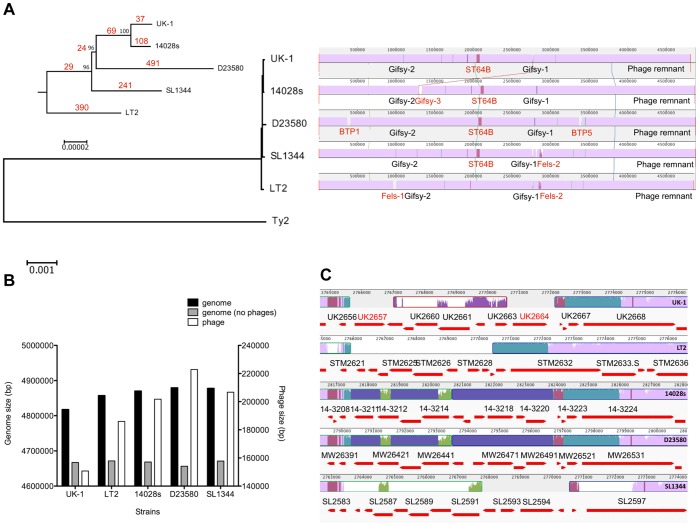
Figure 2. Phylogenetic relationship of the five S. Typhimurium strains.
(A) The phylogenetic tree was inferred with ML method based on the conserved genomic sequences. The S. Typhimurium strains are rooted to S. Typhi Ty2. The upper-left subtree shows the phylogenetic relationship of the five strains in a smaller scale. The relationship was supported by the bootstrapping values shown on the subtree. The distance (marked in red) based on the number of SNPs was also presented on the phylogenetic tree. The right panel shows the complete genome alignment of the five strains generated in MAUVE [93]. The regions conserved among all genomes are colored in purple and the regions conserved among subsets of the genomes are colored differently. If the areas contain sequence elements not aligned, those are marked in white. Regions that are not colored indicate no detectable homology among the five genomes in MAUVE. The distinguished phages and phage remnants are marked on the alignment (black: detected among all of the five strains, red: detected in a subset of strains). (B) Comparison of the lengths of genomes, phages, and genomes excluding phage regions among the five S. Typhimurium strains. Length of phages is displayed on the second Y-axis due to the relatively small value of phages in contrast to the whole genome size. (C) Alignment of the UK-1 Gifsy-1 sequence segment harboring the two UK-1 unique genes with sequences from the other four S. Typhimurium strains. The sequence alignments were generated in MAUVE. The color scheme used for the alignment is described in Fig. 2A. The predicted genes in these regions are shown with red solid arrays. Each gene name is indicated with the strain name (UK indicates UK-1, STM indicates LT2, 14- indicates 14028s, MW indicates D23580, and SL indicates SL1344) followed by its locus number obtained from each of the annotation files. The two UK-1 unique genes are marked in red in the UK-1 genome.