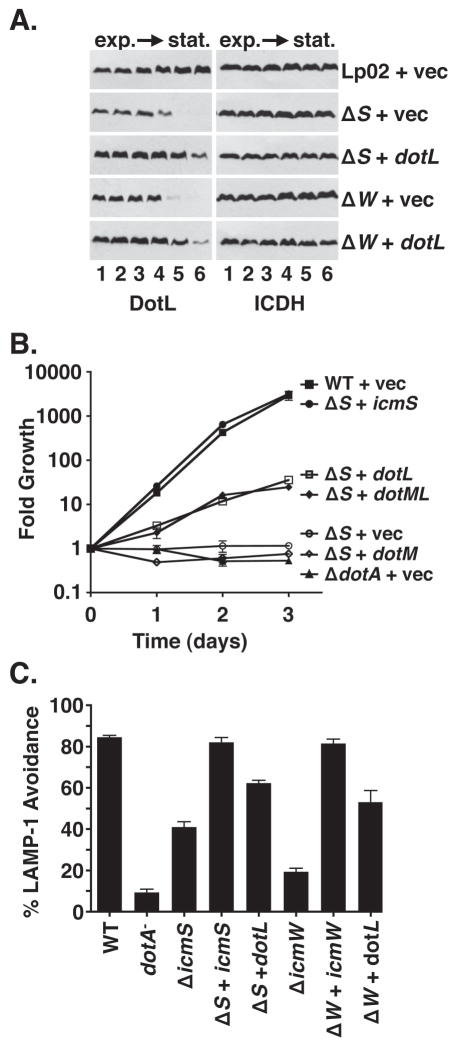
Figure 6. Overexpression of DotL partially suppresses the intracellular growth defects of the ΔicmS and ΔicmW strains.
(A) Destabilization of DotL in the ΔicmS and ΔicmW strains can be partially overcome by overexpression of DotL. L. pneumophila strains were grown in broth and cells were collected at various stages of growth. Whole cell extracts were analyzed by Western blotting using antibodies specific to the proteins listed beneath each panel. Optical densities (600 nm) of the cultures were similar to those described in Fig. 4. Strains were: wild-type plus vector (JV1139), ΔicmS plus vector (JV5156), ΔicmS plus dotL (JV5565), ΔicmW plus vector (JV3658), and ΔicmW plus dotL (JV5570). Western blots for ICDH served as a loading control.
(B) Partial complementation of the intracellular growth defect of the ΔicmS mutant in U937 cells by overexpression of DotL. Strains used were: wild-type plus vector (JV1139, filled squares),ΔicmS plus icmS (JV5157, filled circles), ΔicmS plus dotM + dotL (JV5566, filled diamonds),ΔicmS plus dotL (JV5565, open squares), ΔicmS plus dotM (JV5809, open diamonds) ΔicmS plus vector (JV5156, open circles), and ΔdotA plus vector (JV3029, filled triangles). Fold growth was calculated by dividing the number of colony forming units (CFU) recovered each day by the number of CFU recovered immediately after infection (day 0). Each time point represents the mean and standard deviation of colony forming units (CFU) recovered from triplicate wells. Growth curves are representative of three independent experiments.
(C) Co-localization of L. pneumophila-containing phagosomes with the endocytic marker LAMP-1. Mouse bone marrow-derived macrophages were infected for 1 hour at 37° C, fixed, stained for LAMP-1 and intracellular versus extracellular L. pneumophila, and examined by immunofluoresence microscopy. Strains used were: wild-type plus vector (JV1139), ΔdotA plus vector (JV3029), ΔicmS plus vector (JV5156), ΔicmS plus icmS (JV5157), ΔicmS plus dotL (JV5565), ΔicmW plus vector (JV3658), ΔicmW plus icmW (JV3649), and ΔicmW plus dotL (JV5570). Avoidance of LAMP-1 containing vesicles is shown as the average and standard deviation of four sets of 100 bacteria scored for each strain. Results are representative of two independent experiments.