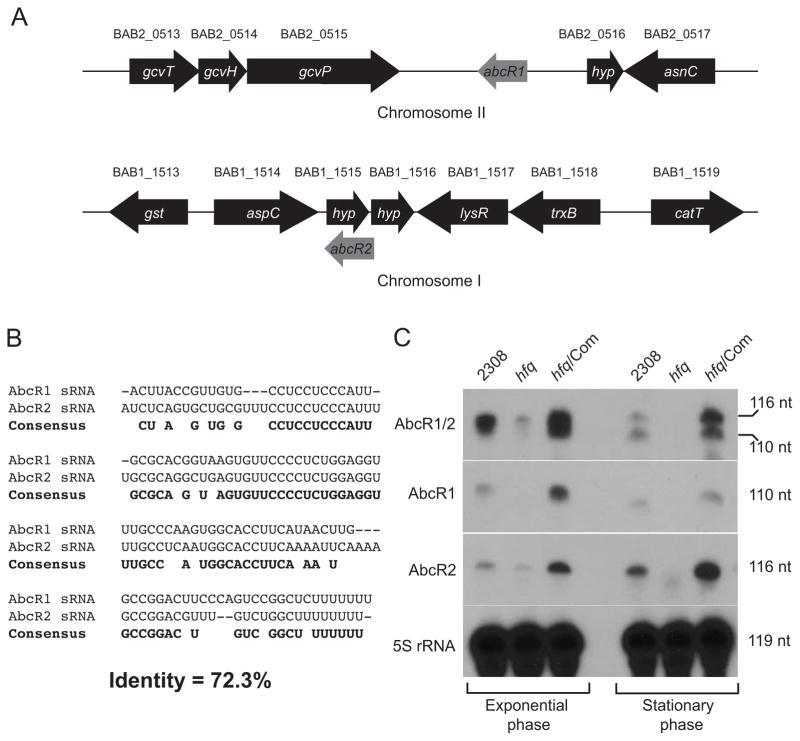
Figure 2. Identification of two small RNAs (sRNAs) in Brucella abortus 2308.
A. Schematic of the loci encoding the AbcR sRNAs.
Due to the high degree of similarity to the A. tumefaciens AbcR sRNAs, the two putative sRNAs in B. abortus 2308 were similarly named AbcR1 and AbcR2. Bioinformatic analyses predicted the B. abortus AbcR1 sRNA to be encoded on chromosome II in the intergenic region between gcvP (BAB2_0515) and a small hypothetical protein-encoding gene (BAB2_0516). The B. abortus AbcR2 sRNA was predicted to be encoded on chromosome I near a small hypothetical protein-encoding gene (BAB1_1515)
B. Alignment of the B. abortus 2308 AbcR sRNAs
The nucleotide sequence of AbcR1 and AbcR2 were aligned using the AlignX® software from Vector NTI® (Invitrogen). Identical nucleotides between the two sRNAs are shown as the consensus sequence in bold font
C.Northern blot detection of AbcR1 and AbcR2
Total RNA isolated from exponential or stationary phase cultures of B. abortus 2308, the hfq mutant strain, and the complemented hfq mutant strain was separated on denaturing polyacrylamide gels. Following transfer to a membrane, a single probe designed to detect both AbcR1 and AbcR2, or probes designed to detect either AbcR1 or AbcR2 were used in the Northern blot analyses. Detection of 5S rRNA was also performed as a loading control. The sizes (in nucleotides) of the detected sRNAs are indicated to the right of the panels.