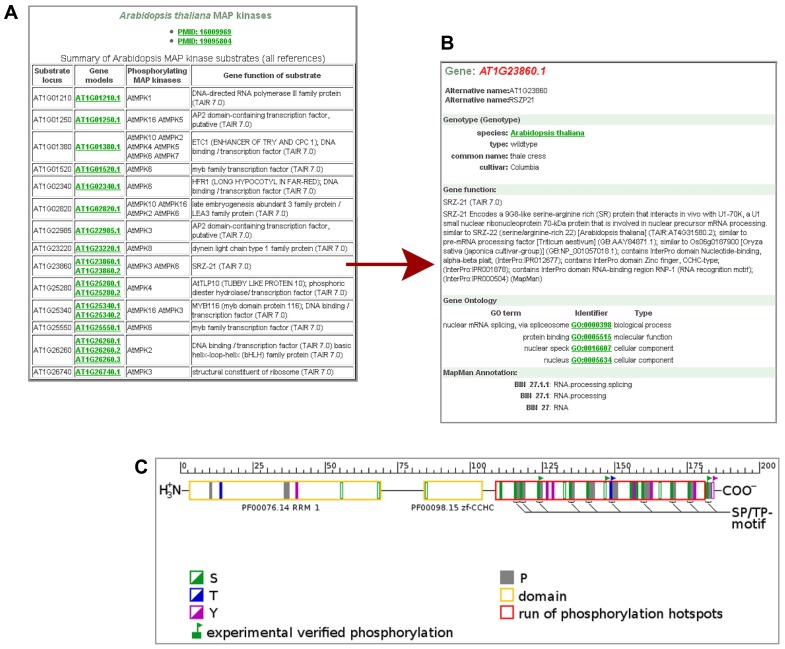
FIGURE 2.
Phosphoproteomic data in GabiPD. (A) List of potential MAP kinase substrates at GabiPD’s Phosphoproteomics page (www.gabipd.org/projects/Arabidopsis_Proteomics/phosphoproteomics_summary.shtml). Substrates were identified by in vitro kinase assays on Arabidopsis protein microarrays. AGI codes of the substrates are linked to the related Gene GreenCard in GabiPD. (B) Gene GreenCard of RSZP21 with integrated kinase assay result. (C) Predicted (filled rectangles in green, blue, and purple) and experimentally verified (flagged rectangles) phosphorylation sites in RSZP21 according to PhosPhAt (Durek et al., 2010; see external links at the Gene GreenCard). The red long box at the C-terminus of the RSZP21 represents a hot spot of phosphorylation predicted recently (Riaño-Pachón et al., 2010). The yellow boxes display conserved protein domains.