Abstract
Fibroproliferative diseases of organs are poorly understood and generally lack effective anti-fibrotic treatments. Our goal was to identify the key regulatory factors in pathologic fibrosis, common between organ-based fibrotic disease. We analyzed 9 microarray datasets publicly available in the GEO datasets from lung, heart, liver and kidney fibrotic disease tissue (489 microarrays total, disease and control). We identified a set of 90 genes differentially expressed in at least five microarray datasets. We used IPA and DAVID analysis to identify gene networks and their molecular functions. A mutual information based network work activity analysis showed that a connective tissue disorders network was the most active for all types of fibrosis included in this analysis. Conclusion: Our analysis indicates that despite different disease manifestation, organ fibrosis share a specific set of genes suggesting the potential for a common origin.
Introduction
Fibrotic diseases are responsible for 45% of deaths1 in the developed world, hence interventions targeting excessive fibrosis are a major therapeutic goal. The concept of 'core' and 'regulatory' pathways in focused on the fundamental regulatory pathways involved in multi-organ fibrosis, specific for human fibrotic disease rather than mouse models, is still an enigma as recently reviewed in Nature Medicine2. Fibrosis is a degenerative process that can lead to end-stage disease and loss of function in the lung3,4, heart5, liver6 or kidney7 (top 15 causes of deaths in U.S.)8. Organ fibrosis is marked by fibroblast activation and abundant extracellular matrix (ECM) deposition, suggesting abnormal wound healing. Fibroblasts are the main contributor to ECM remodeling and excessive collagen deposition in fibrosis. Fibroblast transition to myofibroblast is a common denominator for pathologic fibrosis1 and tumor stroma9–11. In addition, pathological organ fibrosis share several common pathways and biological processes, like TGF-β, MAPK, and PDGF activation; epithelial-to-mesenchymal transition (EMT); metalloproteinase (MMP) activation; mechanical tension; oxidative stress; and inflammation5,6,12–18.
Despite extensive efforts, there are still large gaps in understanding the fundamental molecular pathways in fibrotic diseases across multiple organs. To date, predictive gene signatures have been interrogated for liver or kidney fibrotic disease, but this approach exists more as separate organ-based disease, as opposed for a connected pathological syndrome. For example, a seven-gene19 predictive signature has been outlined for hepatitis C viral infection-induced liver fibrosis. For kidney fibrosis, comparative analysis of four independent microarray datasets generated the “molecular Banff” signature, a 70-genes signature for acute rejection transcript set and a molecular diagnostic approach for early rejection in renal transplant20. Recently, one study using a cross-organ classifier of fibrotic conditions identified a set of markers of human solid organ fibrosis, applicable for renal post-transplant outcome21. Since fibrotic disorders share similar pathological characteristics, including increased collagen deposition and myofibroblast activation, we hypothesized that common genes and pathways regulating these processes are perturbed in fibrosis and we selected gene expression data from fibrosis in multiple organs.
Identifying common differentially expressed genes in multiple datasets is an effective way to increase the power of discovering genes related to key disease phenotypes regardless of different etiology and tissue specificity. Similar methods have been applied on other diseases such as diabetes22, multiple types of cancers23 and neural diseases24. In this study, we extend this method for fibrotic diseases. However, it has become increasingly accepted that for complicated diseases, genes and their protein products act in networks with orchestrated activities. Therefore it is critical to identify the protein-protein interaction (PPI) networks which are involved in the disease development processes. In this study, we use the common differentially expressed genes as “baits” to fish out relevant PPI networks. Starting with these genes, we adopt a commonly used network analysis software Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com) to infer potential relevant PPI networks. The IPA KnowledgeBase has a large collection of known PPI relationships and thus can serve as a adequate resource for our study. The relevance of the networks with individual fibrotic conditions is determined using a mutual information based network activity score25 which has been applied to many diseases studies such as colon cancers 26,27.
To summarize, we developed a translational bioinformatics pipeline for identifying key genes and networks related to fibrosis. We use techniques that have been validated in various diseases such as diabetes22, cancer23, and neural diseases24 for a novel application to identify common pathways underlying fibrosis of different organs. Our analysis provides a novel view to better understanding the common fundamental pathways in organ fibrosis, by using bioinformatics approach for screening genes and networks across multiple organ fibrosis.
Methods
Data selection
The GEO database was searched for the following fibrotic disorders: idiopathic pulmonary fibrosis (IPF), liver cirrhosis, kidney fibrosis, and heart failure (Figure 1). Our search criteria included: 1) both control and disease groups should be present in the datasets, 2) the tissue samples are collected from disease organ (e.g., immortal cell lines and blood are excluded), and 3) at least 5 samples in each group are required for robust statistical analysis. 3 lung datasets, 2 liver, 2 kidney, and 2 heart datasets were downloaded and used for analysis (Table 1).
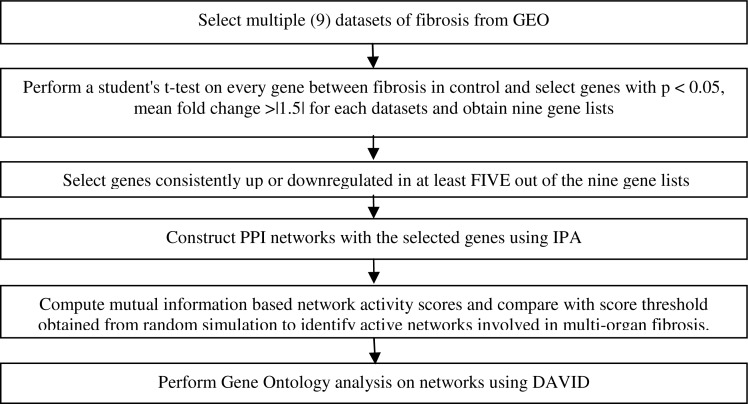
Figure 1.
Workflow for identifying common genes and networks among multiple types of fibrosis.
Table 1:
Components of datasets used for statistical analysis.
| Organ | Datasets | # of Fibrosis | # of Control | Fibrosis |
|---|---|---|---|---|
| Lung | GSE1066728,29 | 31 | 15 | IPF |
| GDS125230,31 | 13 | 11 | IPF | |
| GDS370532 | 7 | 8 | Sarcoidosis | |
| Heart | GSE540633 | 194 | 16 | Heart failure |
| GSE1497534 | 5 | 5 | Atrial fibrillation | |
| Liver | GSE676435 | 13 | 10 | Cirrhosis |
| GSE1432336 | 41 | 19 | Cirrhosis | |
| Kidney | GSE1268237 | 23 | 13 | Tubulointerstitial fibrosis |
| GSE2245938 | 40 | 23 | Interstitial fibrosis/tubular atrophy | |
| Total | 367 | 122 |
Data analysis workflow
Figure 1 gives an overview of our workflow for analyzing genes and PPI networks involved in multiple organ fibrosis. The details of the algorithms are given in following sections.
In general, we first identify genes that are consistently differentially regulated in more than half (5 out of 9) of the datasets. Then PPI networks associated with these genes are established using the Ingenuity Pathway Analysis (IPA) software with the network building function. These networks are further screened for their association with specific fibrotic disease status using a mutual information based network activity score developed by Chuang et al 25. In order to determine the threshold to select gene networks with high mutual information (and hence high relevance with metastasis), 1,000 random sets of genes are selected and the mutual information score is calculated. The threshold is set to be the top five percentile of the random simulations. Finally the selected gene networks with high mutual information are subject to gene set enrichment analysis using DAVID (http://david.abcc.ncifcrf.gov/).
Microarray analysis
All expression values were quantile normalized within each dataset using the MATLAB Bioinformatics Toolbox (R2010a). An unpaired two-sample student's t-test was performed on each gene between fibrosis and control expression values for each dataset. Genes with p < 0.05 and at least 1.5 mean fold change that were consistently upor downregulated were identified as significantly differentially expressed. A list of genes that satisfied these criteria were selected and the number of times each gene was significant in a dataset was counted. Expression values were averaged if multiple probes mapped to the same genes. Probes that did not map to a known gene were eliminated from further analysis. Mean fold change between fibrosis and control was calculated for each gene and averaged across all datasets in which the gene was differentially expressed.
Network analysis using IPA
All genes significant in at least 5 datasets were input into Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com) which has a well curated protein-protein interaction knowledgebase. For this analysis we chose genes that met the criteria for differential expression (>1.5 fold change, p < 0.05) and were found in more than half of the total number of datasets.
Mutual information based network activity analysis
Given a network with a set of genes, a network activity score over different groups of samples was developed previously in 25 based on mutual information (MI). While the details of the method are given in 25, here we outline the steps:
Given a network with N genes across M samples with gene expression levels gij (i = 1, 2, …, N; j = 1,2, …, M), we first transform them into z-scores zij such that for every gene the mean is 0 and standard deviation is 1.
For each sample j, the total z-score zj over all the N genes is .
- Let the sample label vector be c = [c1, c2, …, cM]. For this paper, cj is 0 for non-metastatic patients and is 1 for metastatic patients. The network activity score is defined as
where p(x, y) is the joint probability distribution of the discretized total z-scores and the sample labels and z(′) is the obtained by distributing the total z-scores into [log2 M + 1] evenly spaced levels.
Comparing to other methods such as t- or Wilcox score, MI does not require assumptions on data distribution. In addition, this metric can also accommodate the case when there are subgroups in each group, which is possible among patients with complicated diseases such as fibrosis.
In this study, significant networks identified using IPA were tested using a MATLAB implementation of the mutual information approach applied to all 9 datasets 25.
Random simulation to determine the threshold for network activity scores
Given the network activity scores for all the networks, we need to select the ones with high scores. To find the threshold, we carry out random simulations by randomly select a set of N0 genes where N0 is between 5 and 17 and compute its network activity score. This range of genes was chosen based on the number of genes from our list that IPA used to generate the original networks, called “focus molecules”. This is then repeated 1,000 times, the top five-percentile level is used as the 5% FDR threshold for selecting the networks by Srand. Networks with S > Srand were significantly active in that dataset. Each network had a count corresponding to the number of datasets its S > Srand. Gene ontology information for the genes of the selected networks was accessed using the NIH DAVID webtool.
Results
Differentially expressed gene identification
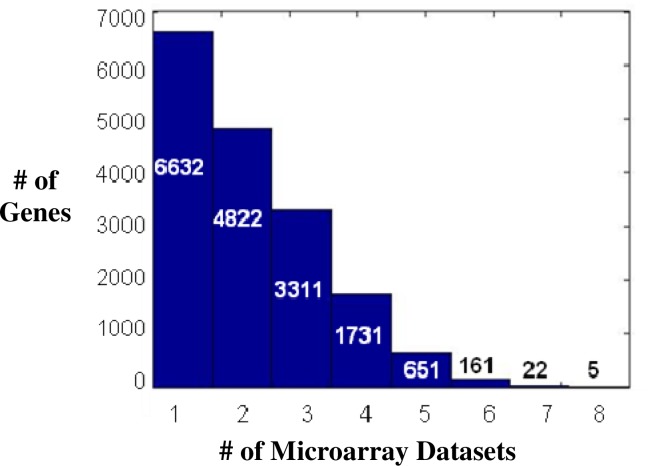
After applying quantile normalization and a t-test for each dataset as described in Methods, the combined list contained 17,335 genes that were expressed in at least one dataset, with p < 0.05 regardless of mean fold change (Figure 2). There were no genes expressed in all 9 datasets with p < 0.05. COL1A1, ITSN1, RUNX3, SMAD2, and WIPF1 were the only genes expressed in 8 out of 9 datasets, with p < 0.05 without considering the fold difference vs. control. These genes have important implications for fibrosis, particularly COL1A139, which encodes the pro-alpha1 chain of type I collagen. SMAD2 is a known to be activated by TGF-β, responsible for the downstream effects of TGF-β like fibroblast activation, myofibroblast production, cell apoptosis and proliferation40,41. ITSN1 (Intersectin 1)42 and WIPF1 (WAS/WASL interacting protein family, member 1)43 are involved in regulating the actin cytoskeleton, are novel for fibrosis. RUNX3 (Runt-related transcription factor 3) is a transcription factor involved in tumor suppression44. WIPF1, an important protein in Wiskott-Aldrich syndrome, was the most frequently differentially expressed gene and met the criteria for 1.5 fold change in 8 datasets. TGFBI (Transforming growth factor, beta-induced) and RNASET2 (Ribonuclease T2) were the only genes differentially expressed in 7 datasets.
Figure 2:
Histogram showing the number of microarray datasets in which a gene shows differential expression (p < 0.05 and mean fold change > 1.5).
Genes in significant networks using mutual information
Of the 17,335 genes expressed in at least one dataset (p < 0.05), only 839 genes were present in 5 or more datasets, over half of the datasets. From these 839 genes, 90 genes were significantly differentially expressed (p < 0.05, |MFC| > 1.5) in at least 5 different datasets (Table 2). Of these, 83 genes were consistently upregulated and 7 genes were consistently down regulated. These 90 genes were input to IPA to uncover the biological functions and pathways involved.
Table 2:
Top 10 genes differentially expressed genes in at least five microarrays with p < 0.05 and |MFC| > 1.5.
| Gene Symbol | Dataset Count | Mean P-value | Mean Fold Change | Molecular Functions |
|---|---|---|---|---|
| MMP7 | 5 | 1.22E-02 | 3.76 | Metalloendopeptidase activity, calcium and zinc ion binding |
| LUM | 5 | 4.45E-03 | 3.60 | Extracellular matrix structural constituent, collagen binding |
| CXCR4 | 5 | 5.26E-03 | 3.51 | Chemokine receptor activity, cytoskeletal protein binding, myosin light chain binding |
| GPNMB | 5 | 1.41E-02 | 3.44 | Binding of: integrin, glycosaminoglycan, heparin, carbohydrate, protein complex |
| ASPN | 5 | 4.98E-03 | 3.20 | Protein binding |
| MT1M | 5 | 2.46E-02 | −2.46 | Copper, zinc and cadmium ion binding |
| AQP3 | 5 | 5.79E-03 | −1.59 | Glycerol transmembrane transporter activity |
| FZD5 | 5 | 2.94E-03 | −1.58 | G-protein coupled receptor activity |
| ST3GAL6 | 5 | 1.21E-02 | −1.58 | Sialyltransferase activity |
| SLC1A1 | 5 | 1.71E-02 | −1.58 | Glutamate:sodium symporter activity |
IPA generated 9 regulatory networks from this list of 90 genes and ignored the 2 genes that were unmapped. Networks 1–7 each contained 35 genes, proteins, other molecules and the regulatory relationships between them, while networks 8 and 9 each contained 3 genes. Networks 8 and 9 were excluded from further analysis due to the small size of the network.
The seven resulting networks shared several genes. Network 5 shared CTSK and the molecule P38 MAPK with Network 1, CCL23 and IL32 with Network 3, and SERPINB3 with Network 6. Network 6 also shared LUM with Network 3 and PLC with Network 7. Networks 4 and 7 had CXCL12 in common.
To test the association of the activity of these networks in the fibrosis datasets, we sought to find the networks that were most relevant to multi-organ fibrosis25. Applying the mutual information approach to the IPA networks returned the activity score (S) for each network, repeated for every dataset (Table 3). It revealed Network 2 as having the highest average score, S = 0.399. A random permutation test of 1000 iterations was performed for each of the 9 datasets, as described in methods. Networks were deemed significantly active for that dataset if S > Srand. Network 1 was significantly active in the greatest number of datasets (4), with the mean of S = 0.3740. Our analysis focused on these two networks because they were most active based on the mutual information scores (Figure 3).
Table 3:
Results of application of mutual information algorithm to each dataset for all seven networks.
| Network | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Srand |
|---|---|---|---|---|---|---|---|---|
| # of Datasets S > Srand | 4 | 3 | 3 | 3 | 1 | 3 | 2 | |
| GSE10667 | 0.4307 | 0.2149 | 0.3213 | 0.2513 | 0.0871 | 0.3300 | 0.1419 | 0.3030 |
| GDS1252 | 0.1855 | 0.0561 | 0.0572 | 0.2969 | 0.4584 | 0.5291 | 0.4967 | 0.4215 |
| GDS3705 | 0.2052 | 0.4131 | 0.3695 | 0.2964 | 0.2267 | 0.6295 | 0.5641 | 0.4458 |
| GSE5406 | 0.1463 | 0.0907 | 0.0700 | 0.0715 | 0.1108 | 0.0357 | 0.0948 | 0.1212 |
| GSE14975 | 0.4490 | 0.3635 | 0.2000 | 0.1145 | 0.2755 | 0.1245 | 0.5145 | 0.7245 |
| GSE12682 | 0.1689 | 0.1443 | 0.0892 | 0.1211 | 0.0846 | 0.0579 | 0.1384 | 0.2371 |
| GSE22459 | 0.1579 | 0.3899 | 0.2717 | 0.3814 | 0.2635 | 0.1972 | 0.1976 | 0.2911 |
| GSE6764 | 0.8365 | 1.0591 | 0.8163 | 1.0519 | 0.4327 | 0.6597 | 0.2936 | 0.7246 |
| GSE14323 | 0.7861 | 0.8674 | 0.7416 | 0.8357 | 0.6618 | 0.6435 | 0.1249 | 0.6790 |
| Average | 0.3740 | 0.3999 | 0.3263 | 0.3801 | 0.2890 | 0.3563 | 0.2852 | 0.4386 |
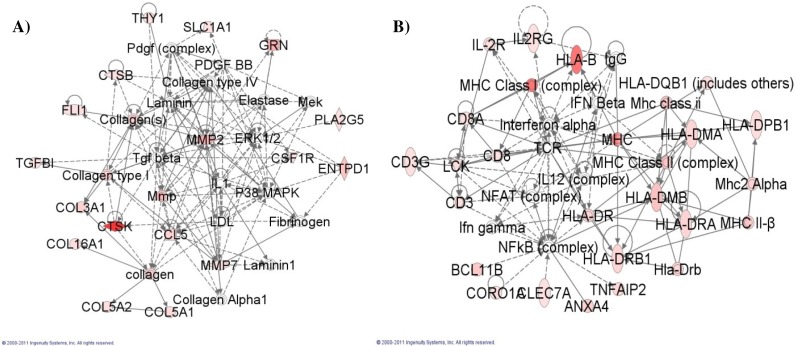
Figure 3:
A) Network graph for Network 1, involved in connective tissue disorders. B) Network graph for Network 2, involved in genetic and skeletal and muscular disorders.
Using DAVID45,46 we identified the molecular functions of the genes belonging to Networks 1 and 2 (Table 4). Proteins and other molecules that were not explicitly genes were ignored by DAVID. DAVID calculated the significance of the molecular functions for the gene list. The molecular functions identified for these genes relate to the top overall functions of their network. The molecular functions of integrin binding and platelet-derived growth factor (PDGF) binding relate the genes of Network 1 to their overall role in connective tissue disorders and tissue development and function. Likewise for Network II, the broad function of genetic disorders could include dysregulation of MHC I binding and MHC II receptor activity.
Table 4:
Top 5 Gene Ontology molecular functions for the components of the most active networks (Networks 1 and 2) using DAVID.
| Network | Components | Top overall network functions | Molecular function of component genes (identified by DAVID) | P-value |
|---|---|---|---|---|
| 1 | CCL5, COL16A1, COL3A1, COL5A1, COL5A2, collagen, Collagen Alpha1, Collagen type I, Collagen type IV, Collagen(s), CSF1R, CTSB, CTSK, Elastase, ENTPD1, ERK1/2, Fibrinogen, FLI1, GRN, IL1, Laminin, Laminin1, LDL, Mek, Mmp, MMP2, MMP7, P38 MAPK, Pdgf (complex), PDGF BB, PLA2G5, SLC1A1, Tgf beta, TGFBI, THY1 | Connective Tissue Disorders, Genetic Disorder, Connective Tissue Development and Function | Integrin binding | 6.7E-7 |
| Protein complex binding | 8.0E-5 | |||
| Extracellular matrix structural constituent | 4.9E-3 | |||
| Endopeptidase activity | 1.0E-2 | |||
| Platelet-derived growth factor binding | 1.3E-2 | |||
| 2 | ANXA4, BCL11B, CD3, CD8, CD3G, CD8A, CLEC7A, CORO1A, HLA-B, HLADMA, HLA-DMB, HLA-DPB1, HLADQB1 (includes others), HLA-DR, HLADRA, Hla-Drb, HLA-DRB1, IFN Beta, Ifn gamma, IgG, IL-2R, IL12 (complex), IL2RG, Interferon alpha, LCK, MHC, MHC Class I (complex), Mhc class ii, MHC Class II (complex), MHC II-β, Mhc2 Alpha, NFAT (complex), NFkB (complex), TCR, TNFAIP2 | Genetic Disorder, Skeletal and Muscular Disorders, Cell-To-Cell Signaling and Interaction | MHC class II receptor activity | 7.5E-12 |
| MHC protein binding | 1.1E-8 | |||
| MHC class I protein binding | 5.5E-7 | |||
| Phosphoinositide 3-kinase binding | 1.1E-2 | |||
| Peptide binding | 2.0E-2 |
Discussion
Since fibrotic disorders are often associated with common syndromes such as chronic inflammation, the goal of our research was to observe foundational gene and network perturbations in multiple types of organ-based fibrosis. The bioinformatic analysis of microarray datasets publicly available for lung, heart, liver and kidney fibrosis has provided an opportunity to investigate the hypothesis that there are foundational molecular mechanisms for all types of organ-based fibrosis. This approach is vital to understanding the core genes involved in fibrosis that mark potential targets for therapy.
The results paint a picture of the development of fibrosis characterized by a core set of genes and molecular pathways. ∼17,000 genes were identified as being expressed in at least one dataset with p < 0.05, but only 0.52% of these genes (83 genes up-regulated and 7 genes down-regulated) were significantly differentially expressed across these datasets and were used for further analysis.
Numerous observations indicate that fibrosis, aging and abnormal wound healing may be linked, as both may represent loss of cellular reserve pathways47,48. This would explain the abnormal expression of genes implicated in stem cells biology, wound repair and epithelial damage beside the known “myofibroblasts” genes.
To understand the function of these core genes, we used two different programs to analyze the genes and identify their biological and molecular functions. First, IPA software provided the top biological functions and revealed that dermatological diseases as the most disorder that aligned the most significant fit for genes found differentially expressed at p < 10−16. Other relevant functions identified by this approach included inflammatory disease, connective tissue disorders, genetic disorder, and respiratory and cardiac disease, all with p-values < 10−7. IPA then generated networks using the list of genes as “seed” for these networks, with the activity of these networks scored by the mutual information algorithm and confirmed by subjecting each network to a test of significance against a random simulation of mutual information. This aided in identifying which processes were most perturbed by fibrosis. The network deemed the most active in the greatest number of networks (Network 1) and the network most active overall with the highest average mutual information score (Network 2) are involved in critical cellular, immune and matrix functions. The genes and molecules of Network 1 play a key role in connective tissue disorders. Network 2 is important in genetic, skeletal and muscular disorders through several HLA and MHC genes involved in antigen presentation and recognition in immune system response. Secondly, DAVID analysis of the selected genes identified molecular functions of these genes that are in line with the biological functions of their networks and the pathogenesis of fibrosis. Network 1 contained genes important in cell-matrix interactions like integrins are significantly dysregulated per our analysis of the microarray datasets and are potentially play important roles in fibrosis.
In addition, there were many genes with p < 0.05 that were expressed in 5 or more datasets, but had mean fold-changes of smaller magnitude than 1.5 (data not shown). These genes are members of pathways not directly involved in fibrosis, and represent other potential targets. The top biological functions of these networks were involved in hematopoiesis, tissue morphology, cell cycle and death, and may indicate the important influence of stem cells, developmental pathways and repair on organ fibrosis. These functions has p values ranging from 10−5 to 10−2, less significant that the functions of the differentially expressed genes.
Our analysis revealed several genes previously unknown for fibrosis and widely up-regulated in multi-organ fibrosis: WIPF1 (the highest expression, 8 microarrys), ITGBL1 (Integrin, beta-like 1 (with EGF-like repeat domains)) involved in inflammation49, EPHA3 (EPH receptor A3) in mediating developmental processes and homing of hematopoietic cells50,51, GRN (Granulin) in epithelial development, tissue remodeling and wound healing52,53. Most of the common down-regulated genes present in our analysis are novel for organ fibrosis: MT1M (Metallothionein 1M) linked to progressive degeneration of motoneurons in sporadic amyotrophic lateral sclerosis54, FZD5 (Frizzled family receptor 5) which is part of WNT signaling and ST3GAL6 (ST3 beta-galactoside alpha-2,3-sialyltransferase 6) important for stem cell development and regeneration55,56, SLC1A1 (Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1), a major epithelial transporter of glutamate and aspartate57. Chronic inflammation, extracellular matrix remodeling and epithelial development as part of abnormal wound healing involved in fibrosis are well represented in our analysis by up-regulated genes like LUM (Lumican)58, GPNMB (Glycoprotein (transmembrane) nmb59) or genes downregulated: AQP3 (Aquaporin 3 (Gill blood group))60,61 and IL6R (Interleukin 6 receptor)62,63. It was interesting to see that our analysis has identified a set of well known genes for fibrosis such as chemokine CXCR4 ((C-X-C motif) receptor 4), SDF1/CXCL12 (Chemokine (C-X-C motif) ligand 12), metalloproteinases (MMP7, MMP2, at the level of multi-organ fibrosis. In addition, several genes from our analysis have been previously described in gene signatures for organ fibrosis. 14 genes from our list are present in the molecular Banff signature for acute kidney transplant rejection20. In another fibrosis study, two genes from our list, MMP2 and COL3A1, were found in fibrotic heart, kidney, lung and pancreas tissues, while ADAM28 was found only in lung tissues21.
Our analysis indicates that besides regular fibroblasts-myofibroblast genes, there is a common set of genes abnormally expressed, involved in epithelial development, stem cells regeneration and inflammation. We go beyond the approach of single organ fibrosis to address core genes and molecules involved in fibrosis that are present in multiple organs. While these genes may not be the original disease causing genes whose genetic variations leading to the onset or predisposition of these diseases, they reflect a set of potential commonly changed phenotypes at the gene transcription level which can be experimentally tested and as well as targeted in therapy. Nevertheless, our results are only the beginning, as the genes identified give way to experimental research to confirm the role of the identified genes in multi-organ fibrosis, and identify therapeutic targets to slow or even reverse fibrotic activity.
References
- 1.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008 Jan;214(2):199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehal WZ, Iredale J, Friedman SL. Scraping fibrosis: Expressway to the core of fibrosis. Nat Med. 2011 May;17(5):552–3. doi: 10.1038/nm0511-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.du Bois RM. Strategies for treating idiopathic pulmonary fibrosis. Nat Rev Drug Discov. 2010 Feb;9(2):129–40. doi: 10.1038/nrd2958. [DOI] [PubMed] [Google Scholar]
- 4.Hubbard RB, Smith C, Le Jeune I, et al. The association between idiopathic pulmonary fibrosis and vascular disease: a population-based study. Am J Respir Crit Care Med. 2008 Dec 15;178(12):1257–61. doi: 10.1164/rccm.200805-725OC. [DOI] [PubMed] [Google Scholar]
- 5.van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J. Myocardial remodeling after infarction: the role of myofibroblasts. Nat Rev Cardiol. 2010 Jan;7(1):30–7. doi: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- 6.Friedman SL. Evolving challenges in hepatic fibrosis. Nat Rev Gastroenterol Hepatol. 2010 Aug;7(8):425–36. doi: 10.1038/nrgastro.2010.97. [DOI] [PubMed] [Google Scholar]
- 7.Boor P, Ostendorf T, Floege J. Renal fibrosis: novel insights into mechanisms and therapeutic targets. Nat Rev Nephrol. 2010 Nov;6(11):643–56. doi: 10.1038/nrneph.2010.120. [DOI] [PubMed] [Google Scholar]
- 8.Heron M, Hoyert DL, Murphy SL, Xu J, Kochanek KD, Tejada-Vera B. Deaths: final data for 2006. Natl Vital Stat Rep. 2009 Apr 17;57(14):1–134. [PubMed] [Google Scholar]
- 9.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006 May;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 10.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005 Feb;15(1):97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004 Nov 18;432(7015):332–7. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minagawa S, Araya J, Numata T, Nojiri S, Hara H, Yumino Y, et al. Accelerated epithelial cell senescence in IPF and the inhibitory role of SIRT6 in TGF-beta-induced senescence of human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2011 Mar;300(3):L391–401. doi: 10.1152/ajplung.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino M, Tosoni A, Nebuloni M. Direct contribution of epithelium to organ fibrosis: epithelialmesenchymal transition. Hum Pathol. 2009 Oct;40(10):1365–76. doi: 10.1016/j.humpath.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Turner NA, Warburton P, O'Regan DJ, Ball SG, Porter KE. Modulatory effect of interleukin-1alpha on expression of structural matrix proteins, MMPs and TIMPs in human cardiac myofibroblasts: role of p38 MAP kinase. Matrix Biol. 2010 Sep;29(7):613–20. doi: 10.1016/j.matbio.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson O, Diebold D, Fan D, Peterson M, Nho RS, Bitterman PB, et al. Fibrotic myofibroblasts manifest genome-wide derangements of translational control. PLoS One. 2008;3(9):e3220. doi: 10.1371/journal.pone.0003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neilson EG. Mechanisms of disease: Fibroblasts--a new look at an old problem. Nat Clin Pract Nephrol. 2006 Feb;2(2):101–8. doi: 10.1038/ncpneph0093. [DOI] [PubMed] [Google Scholar]
- 17.Ludwicka-Bradley A, Silver RM, Bogatkevich GS. Coagulation and Autoimmunity in Scleroderma Interstitial Lung Disease. Semin Arthritis Rheum. 2010 Dec 16; doi: 10.1016/j.semarthrit.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyton SR, Ghajar CM, Khatiwala CB, Putnam AJ. The emergence of ECM mechanics and cytoskeletal tension as important regulators of cell function. Cell Biochem Biophys. 2007;47(2):300–20. doi: 10.1007/s12013-007-0004-y. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Shiffman ML, Friedman S, Venkatesh R, Bzowej N, Abar OT, et al. A 7 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis C. Hepatology. 2007 Aug;46(2):297–306. doi: 10.1002/hep.21695. [DOI] [PubMed] [Google Scholar]
- 20.Saint-Mezard P, Berthier CC, Zhang H, Hertig A, Kaiser S, Schumacher M, et al. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009 Mar;22(3):293–302. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 21.Rodder S, Scherer A, Korner M, Marti HP. A subset of metzincins and related genes constitutes a marker of human solid organ fibrosis. Virchows Arch. 2011 Apr;458(4):487–96. doi: 10.1007/s00428-011-1052-7. [DOI] [PubMed] [Google Scholar]
- 22.English SB, Butte AJ. Evaluation and integration of 49 genome-wide experiments and the prediction of previously unknown obesity-related genes. Bioinformatics (Oxford, England) 2007 Nov 1;23(21):2910–7. doi: 10.1093/bioinformatics/btm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rybaczyk LA, Bashaw MJ, Pathak DR, Huang K. An indicator of cancer: downregulation of monoamine oxidase-A in multiple organs and species. BMC genomics. 2008;9:134. doi: 10.1186/1471-2164-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseveleki V, Rubio R, Vamvakas SS, White J, Taoufik E, Petit E, et al. Comparative gene expression analysis in mouse models for multiple sclerosis, Alzheimer's disease and stroke for identifying commonly regulated and disease-specific gene changes. Genomics. Aug;96(2):82–91. doi: 10.1016/j.ygeno.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nibbe RK, Chowdhury SA, Koyuturk M, Ewing R, Chance MR. Protein-protein interaction networks and subnetworks in the biology of disease. Wiley interdisciplinary reviews. May-Jun;3(3):357–67. doi: 10.1002/wsbm.121. [DOI] [PubMed] [Google Scholar]
- 27.Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009 Apr;8(4):827–45. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009 Jul 15;180(2):167–75. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, Lokshin AE, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008 Apr 29;5(4):e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005 Sep;2(9):e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med. 2006 Dec 25;203(13):2895–906. doi: 10.1084/jem.20061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockstone HE, Sanderson S, Kulakova N, Baban D, Leonard A, Kok WL, et al. Gene set analysis of lung samples provides insight into pathogenesis of progressive, fibrotic pulmonary sarcoidosis. Am J Respir Crit Care Med. 2010 Jun 15;181(12):1367–75. doi: 10.1164/rccm.200912-1855OC. [DOI] [PubMed] [Google Scholar]
- 33.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, et al. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006 Sep 19;114(12):1269–76. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 34.Adam O, Lavall D, Theobald K, Hohl M, Grube M, Ameling S, et al. Rac1-induced connective tissue growth factor regulates connexin 43 and N-cadherin expression in atrial fibrillation. J Am Coll Cardiol. 2010 Feb 2;55(5):469–80. doi: 10.1016/j.jacc.2009.08.064. [DOI] [PubMed] [Google Scholar]
- 35.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007 Apr;45(4):938–47. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 36.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009 Mar-Apr;15(3–4):85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si H, Banga RS, Kapitsinou P, Ramaiah M, Lawrence J, Kambhampati G, et al. Human and murine kidneys show gender- and species-specific gene expression differences in response to injury. PLoS One. 2009;4(3):e4802. doi: 10.1371/journal.pone.0004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010 Nov;21(11):1987–97. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutroneo KR. TGF-beta-induced fibrosis and SMAD signaling: oligo decoys as natural therapeutics for inhibition of tissue fibrosis and scarring. Wound Repair Regen. 2007 Sep-Oct;15(Suppl 1):S54–60. doi: 10.1111/j.1524-475X.2007.00226.x. [DOI] [PubMed] [Google Scholar]
- 40.Ikushima H, Miyazono K. Biology of Transforming Growth Factor-beta Signalin. Curr Pharm Biotechnol. 2011 May 27; doi: 10.2174/138920111798808419. [DOI] [PubMed] [Google Scholar]
- 41.Santibanez JF, Quintanilla M, Bernabeu C. TGF-beta/TGF-beta receptor system and its role in physiological and pathological conditions. Clin Sci (Lond) 2011 Sep 1;121(6):233–51. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- 42.O'Bryan JP. Intersecting pathways in cell biology. Sci Signal. 2010;3(152):re10. doi: 10.1126/scisignal.3152re10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anton IM, Jones GE. WIP: a multifunctional protein involved in actin cytoskeleton regulation. Eur J Cell Biol. 2006 Apr;(3–4):85. 295–304. doi: 10.1016/j.ejcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010 May 6;29(18):2605–15. doi: 10.1038/onc.2010.88. [DOI] [PubMed] [Google Scholar]
- 45.Wang M, Munier F, Araki-Saski K, Schorderet D. TGFBI gene transcript is transforming growth factor-beta1-responsive and cell density-dependent in a human corneal epithelial cell line. Ophthalmic Genet. 2002 Dec;23(4):237–45. doi: 10.1076/opge.23.4.237.13884. [DOI] [PubMed] [Google Scholar]
- 46.Luhtala N, Parker R. T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci. 2010 May;35(5):253–9. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 Jan;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009 Aug;58(8):1078–83. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- 50.Guan M, Liu L, Zhao X, Wu Q, Yu B, Shao Y, et al. Copy number variations of EphA3 are associated with multiple types of hematologic malignancies. Clin Lymphoma Myeloma Leuk. 2011 Feb 1;11(1):50–3. doi: 10.3816/CLML.2011.n.006. [DOI] [PubMed] [Google Scholar]
- 51.Ting MJ, Day BW, Spanevello MD, Boyd AW. Activation of ephrin A proteins influences hematopoietic stem cell adhesion and trafficking patterns. Exp Hematol. 2010 Nov;38(11):1087–98. doi: 10.1016/j.exphem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Davidson B, Alejandro E, Florenes VA, Goderstad JM, Risberg B, Kristensen GB, et al. Granulin-epithelin precursor is a novel prognostic marker in epithelial ovarian carcinoma. Cancer. 2004 May 15;100(10):2139–47. doi: 10.1002/cncr.20219. [DOI] [PubMed] [Google Scholar]
- 53.Bateman A, Bennett HP. The granulin gene family: from cancer to dementia. Bioessays. 2009 Nov;31(11):1245–54. doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 54.Lederer CW, Torrisi A, Pantelidou M, Santama N, Cavallaro S. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics. 2007;8:26. doi: 10.1186/1471-2164-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh M. WNT signaling in stem cell biology and regenerative medicine. Curr Drug Targets. 2008 Jul;9(7):565–70. doi: 10.2174/138945008784911750. [DOI] [PubMed] [Google Scholar]
- 56.Hemmoranta H, Satomaa T, Blomqvist M, Heiskanen A, Aitio O, Saarinen J, et al. N-glycan structures and associated gene expression reflect the characteristic N-glycosylation pattern of human hematopoietic stem and progenitor cells. Exp Hematol. 2007 Aug;35(8):1279–92. doi: 10.1016/j.exphem.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Bailey CG, Ryan RM, Thoeng AD, Ng C, King K, Vanslambrouck JM, et al. Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. J Clin Invest. 2011 Jan 4;121(1):446–53. doi: 10.1172/JCI44474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu F, Chakravarti S. Differential expression of inflammatory and fibrogenic genes and their regulation by NF-kappaB inhibition in a mouse model of chronic colitis. J Immunol. 2007 Nov 15;179(10):6988–7000. doi: 10.4049/jimmunol.179.10.6988. [DOI] [PubMed] [Google Scholar]
- 59.Psarras S, Mavroidis M, Sanoudou D, Davos CH, Xanthou G, Varela AE, et al. Regulation of adverse remodelling by osteopontin in a genetic heart failure model. Eur Heart J. 2011 Jul 20; doi: 10.1093/eurheartj/ehr119. [DOI] [PubMed] [Google Scholar]
- 60.Kreda SM, Gynn MC, Fenstermacher DA, Boucher RC, Gabriel SE. Expression and localization of epithelial aquaporins in the adult human lung. Am J Respir Cell Mol Biol. 2001 Mar;24(3):224–34. doi: 10.1165/ajrcmb.24.3.4367. [DOI] [PubMed] [Google Scholar]
- 61.Bedford JJ, Leader JP, Walker RJ. Aquaporin expression in normal human kidney and in renal disease. J Am Soc Nephrol. 2003 Oct;14(10):2581–7. doi: 10.1097/01.asn.0000089566.28106.f6. [DOI] [PubMed] [Google Scholar]
- 62.McGreal EP, Davies PL, Powell W, Rose-John S, Spiller OB, Doull I, et al. Inactivation of IL-6 and soluble IL-6 receptor by neutrophil derived serine proteases in cystic fibrosis. Biochim Biophys Acta. 2010 Jul-Aug;1802(7–8):649–58. doi: 10.1016/j.bbadis.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Nechemia-Arbely Y, Barkan D, Pizov G, Shriki A, Rose-John S, Galun E, et al. IL-6/IL-6R axis plays a critical role in acute kidney injury. J Am Soc Nephrol. 2008 Jun;19(6):1106–15. doi: 10.1681/ASN.2007070744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin P, Parkhurst SM. Parallels between tissue repair and embryo morphogenesis. Development. 2004 Jul;131(13):3021–34. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- 65.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med. 2008 Mar 4;5(3):e62. doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]