Abstract
Negation of clinical named entities is common in clinical documents and is a crucial factor to accurately compile patients’ clinical conditions and to further support complex phenotype detection. In 2009, Mayo Clinic released the clinical Text Analysis and Knowledge Extraction System (cTAKES), which includes a negation annotator that identifies negation status of a named entity by searching for negation words within a fixed word distance. However, this negation strategy is not sophisticated enough to correctly identify complicated patterns of negation. This paper aims to investigate whether the dependency structure from the cTAKES dependency parser can improve the negation detection performance. Manually compiled negation rules, derived from dependency paths were tested. Dependency negation rules do not limit the negation scope to word distance; instead, they are based on syntactic context. We found that using a dependency-based negation proved a superior alternative to the current cTAKES negation annotator.
Introduction
In clinical documents, named entities often include diseases and disorders or signs and symptoms. Retrieving these clinical named entities is important for both clinical research and patient health care. However, not all clinical named entities in clinical documents are actually present in patients. Those mentioned but not present are often ruled out or absent [1]. Thus, it is critical to differentiate negative named entities from positive ones in order to accurately compile patients’ clinical conditions and produce patient profiles that are fundamental for high throughput phenotyping and other modes of secondary use.
Mayo Clinic developed the clinical Text Analysis and Knowledge Extraction System (cTAKES) and first released it to the open source community1 in 2009. cTAKES is a comprehensive, modular, extensible information extraction system, which processes clinical narratives and identifies clinical named entities along with attributes; one of these attributes is negation [2, 3]. cTAKES has been applied to various use cases in clinical domains [4–7].
The original cTAKES negation module is roughly based on NegEx [8], which implements a regular expression pattern matching algorithm to search for manually defined negation words around named entities, within a fixed distance of word tokens. cTAKES determines negation by searching negation keywords within seven words on the left and the right of the named entity. This simple algorithm catches most conditions explicitly negated in clinical narratives. However, a fixed negation scope is not flexible enough to deal with complicated negation mentions, and cannot correctly identify negation when contextual understanding is necessary. One way to better understand contexts and handle difficult negations is to use deeper syntactic structure, but this has largely been ignored in clinical natural language processing. Mayo Clinic’s latest release of the cTAKES (version 1.1.0) addresses this historical gap by introducing a dependency parser module that provides the rich syntactic information.
In this paper, we investigated the degree of enhancement in negation detection attributable to a dependency structure. We have tested the negation accuracy of our dependency parser-based negation (DepNeg) against the baseline of word distance-based cTAKES negation, and found that DepNeg performed better.
Background
Negation in clinical narratives has been investigated in numerous ways. Many negation algorithms, including the existing cTAKES negation module, take a rule-based approach, with a variety of techniques: regular expression pattern matching [8], lexical scan with context free grammar [9], or invoking a negation ontology [10]. Some negation algorithms use machine learning techniques, such as naïve Bayes and decision trees [11]. In comparison with the proposed work, these approaches ignore or make marginal use of deeper syntactic structure. Similar work to ours has invoked syntactic information using grammatical parsing [12], but our approach differs in that we use dependency parses which directly encode thematic roles like subject and object instead of using explicit grammatical phrasal structure.
Previous writers suggest that negation in clinical narratives is relatively uncomplicated [9, 13] and that a limited set of negation words cover a large portion of negation mentions [1]. With that hypothesis, a rule based approach that uses a lexical pattern matching algorithm has been widely applied. However, properly defining negation scope is a challenging task for complicated negation mentions. The rich syntactic and relation information from dependency parses could aid in mitigating this issue. In the next section, we begin with a brief background of dependency parsing, focusing on the output parses in a graphical and hierarchical representation. In the method section we explain how dependency path rules can be applied in negation.
Dependency Parsing
Figure 1 shows an example of an output dependency parse for the sentence “There was no evidence of edema.” Dependency parsing is typically defined at the sentence level.
Figure 1.
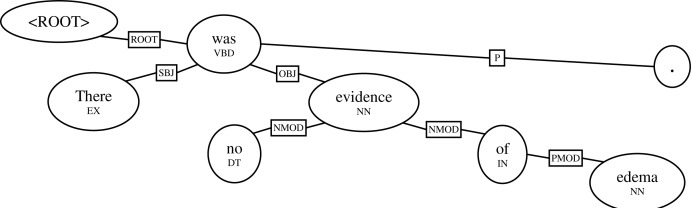
An example dependency parse.
Unlike deep parses, dependency parses do not explicitly represent phrasal structure like noun phrases or verb phrases. Rather, syntactic structure is expressed as directed dependencies between words, so that words (ovals) higher on the chart ‘generate’ words lower on the chart. An additional root node, with only one dependent (in this case, “was”) is present for every dependency parse.
The dependencies may be labeled with relations (boxes in the diagram), such as the subject relation SBJ, the object relation OBJ, a noun-modifying relation NMOD, a preposition-modifying relation PMOD, or a punctuation mark P. Additional information may be included with each word, such as the part of speech (POS) that has been included underneath each word in the example.
The goal of a dependency parser is to induce this kind of structure for a given utterance – it is typically required that the sentence be tokenized, POS-tagged, and lemmatized. Two main approaches have been explored: graph-based approaches and transition-based approaches. cTAKES invokes the ClearParser [14, 15] dependency parser, which takes the latter approach – it proceeds in a single pass over the words in the sentence, determining what dependencies are present after each input word. ClearParser improves on previous transition-based approaches by adding an extra transition that substantially speeds up computation, making it approximate a linear execution time. These speed characteristics, coupled with state-of-the-art accuracy, make it an arguably better choice for large-scale processing than other options [15] such as MaltParser [16] or the Stanford Dependency Parser [17].
Data
To develop dependency patterns rules for negation, we examined 41 sentences from the 2010 I2B2/VA NLP challenge data2 that contained an assertion status of “absent.” The test data consists of 160 Mayo clinical notes in which named entities and their negation were manually annotated by four domain experts. Inter-annotator agreement (IAA) has been examined and reported in Table 1 [2].
Table 1.
Inter-annotator agreement for negation on the 160-note Mayo corpus.
A12 is the gold standard created by A1 and A2; A34 is the gold standard created by A3 and A4
| Annotation sets | IAA |
|---|---|
| A1, A2, A3, A4 | 0.790 |
| A12, A34 | 0.848 |
IAA = matches / (matches + non-matches)
Our final “consensus” data was derived after an adjudication stage during which all disagreements were discussed and a decision on the final label was reached. This final “consensus” data was used in the test experimentation.
Methods
This study applied dependency parses to negation detection. Given a “focal” named entity in clinical text, we seek to determine whether that named entity is negated. To do so with dependency parses, we first examined a development (i.e., training) set of sentences from the I2B2/VA NLP challenge and manually designed syntactic rules which indicated when terms were negated. We then compared our dependency parser-based approach against the existing cTAKES negation module.
The cTAKES pipeline performs standard text pre-processing on clinical text (Figure 2, refer to [2] for details), and satisfies the pre-conditions for a dependency parser. Recall that dependency parsing is defined on pre-tokenized sentences, typically with POS tags and lemmas. cTAKES includes a lemmatizer based on the LVG Specialist Lexicon3 and the Maximum Entropy-based POS tagger from OpenNLP4. Thus, in this pipeline, a dependency parser and candidate named entity recognition (NER), such as signs/symptoms and diseases/disorders, (both in bold in Figure 2) are available to the negation detection module.
Figure 2.

A block diagram of modules in the cTAKES pipeline.
We based negation rules on the dependency path between the focus (i.e., named entity) and each other term (i.e., hypothesized negation words) in the text. Dependency paths can easily be calculated between any two nodes on a dependency tree. In Figure 3, “edema” is a focus term discovered by the dictionary lookup module, and the dependency path between “no” and “edema” is simply a subset of the dependency parse.
Figure 3.
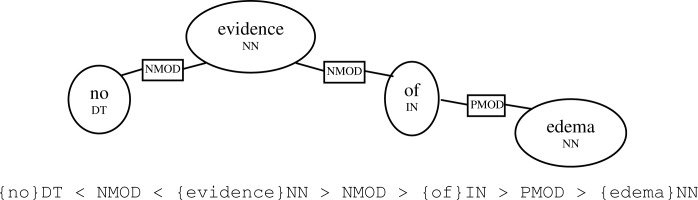
An example of a dependency path.
This dependency path can also be represented as a string which enables path matching implementation. In this paper, however, we will use the graphical representation for clarity.
To use the cTAKES named entities as focus annotations for DepNeg, we followed the steps below.
Find the head node. For each focus annotation, we first normalized multi-word foci to the highest node or headword to represent the whole span, since dependency paths are undefined on multi-word expressions. For example, if “rib fracture” was the focus annotation in “No evidence of rib fracture,” we would choose “fracture” as the representative word (assuming “rib” is a dependent of “fracture”).
Calculate paths. Given the representative word of the named entity, we considered each other token in the sentence, and calculated its path to the focus annotation.
Match paths. For each token-to-focus dependency path, evaluate whether the dependency path matches a pattern in a bank of acceptable dependency path patterns (see below). If there is any match, negate the named entity.
Dependency Path Patterns
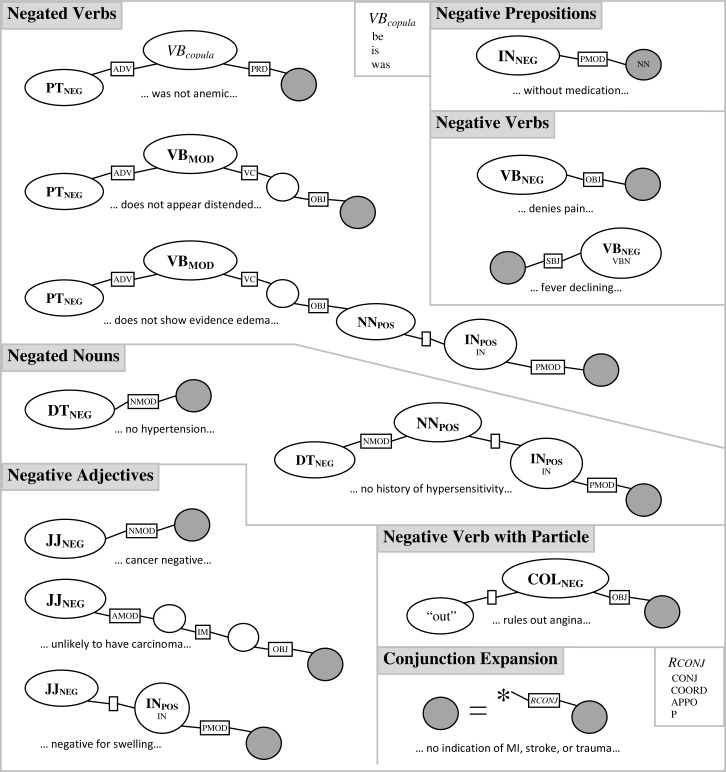
The dependency path patterns (Figure 4) were developed by examining a small subset of data from the 2010 I2B2/VA NLP challenge. The I2B2 data were run through the whole cTAKES pipeline, including the dependency parser. Figure 4 divides the negation patterns on dependency paths up into several types of syntactic constructions which can result in the negation of a named entity.
Negated Verbs (3 patterns). Negating a verb (e.g., “did not show…”) usually implies that the whole verb phrase is negated, and thus that objects or complements should be negated.
Negative Verbs (3 patterns). Some verbs (e.g., “denies”) inherently disqualify the direct object of the verb. A special case also exists, in which the verb is only negative if a particle is present (e.g., “rules out”).
Negative Prepositions (1 pattern). A few prepositions (e.g., “without”) will negate the object of the preposition.
Negated Nouns (2 patterns). Certain negating determiners (e.g., “no”) will negate the nouns they modify.
Negative Adjectives (3 patterns). Some adjectives (e.g., “negative”) will negate the nouns they modify.
Conjunction Expansion. This rule is in fact a meta-rule, allowing for conjunctions or lists of any kind to be used in every other rule.
Figure 4.
DepNeg negation patterns based on dependency paths, with examples
These pattern rules were implemented in regular expression form on the string representation of paths, but are presented in graphical form in Figure 4. The graphical patterns are similar to the notation we have used for dependency trees and dependency paths. One difference is that bold or italicized text represents a set of terms (cross-indexed with Figure 5); acceptable paths would then draw from that set. Additionally, blank nodes or relations indicate that any term or relationship would be acceptable, given that the other conditions were satisfied.
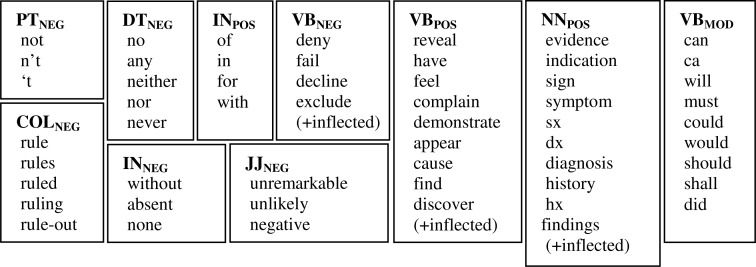
Figure 5.
cTAKES negation module keywords, reused in DepNeg
The existing cTAKES negation module uses negation words and other keywords in its term pattern matching (see Figure 5). To isolate the contribution of dependency parses to negation, we used the same sets of negation words and keywords in the DepNeg dependency path patterns. It should be clarified that while we have motivated the rules by their syntactic behavior, negation words and other keywords used in the existing cTAKES negation module are not entirely consistent or optimal for a dependency-based negation paradigm. Thus the syntactic categories or POS tags may at times be misnomers – INNEG, for example, should be the set of negative prepositions, but it includes words such as “absent” and “none” which are not prepositions.
Evaluation Measurement
For evaluation of system outputs we used:
Where TP is the number of true positives, FP is the number of false positives, TN is the number of true negatives, and FN is the number of false negatives.
Results
Recall that to achieve a fair comparison of negation performance, we used the same set of negation indication words in both DepNeg and cTAKES negation. However, DepNeg did not use pseudo-negation phrases (i.e., double negation, for example, “not rule out”) which was implemented in cTAKES negation.
We used the dependency parser in the latest version of cTAKES (version 1.1.0) to determine a dependency structure between clinical named entities and negation indication words. The dependency parser was trained on the merged data of three sets, clinical questions5, GENIA [18], and Wall Street Journal (WSJ) [19].
Table 2 shows negation statistics and evaluation of the original cTAKES negation (Table 2 (a)) and the DepNeg (Table 2 (b)) model on the test set (Mayo’s 160 clinical notes). Negation performance of two clinical named entities was reported. Signs/symptoms contain 111 negated entities (85 TPs + 26 FNs) out of total 381 entities and diseases/disorders contain 123 negated entities (102 TPs + 21 FNs) out of total 854 entities. In other words, approximately 30% of signs/symptoms and 10% of diseases/disorders are negated in the actual data.
Table 2.
Negation statistics and evaluation on the test set
| (a) cTAKES Negation | ||||||||
|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | precision | recall | F-score | accuracy | |
| Signs/symptoms | 85 | 258 | 12 | 26 | 0.876 | 0.766 | 0.817 | 0.900 |
| Diseases/disorders | 102 | 709 | 22 | 21 | 0.823 | 0.829 | 0.826 | 0.950 |
| All | 187 | 967 | 34 | 47 | 0.846 | 0.799 | 0.822† | 0.934† |
| (b) DepNeg | ||||||||
|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | precision | recall | F-score | accuracy | |
| Signs/symptoms | 85 | 269 | 1 | 26 | 0.988 | 0.766 | 0.863 | 0.929 |
| Diseases/disorders | 88 | 726 | 5 | 35 | 0.946 | 0.715 | 0.815 | 0.953 |
| All | 173 | 995 | 6 | 61 | 0.966 | 0.739 | 0.838† | 0.946† |
micro average – i.e., obtained by using a global count of each named entity and averaging these sums.
Overall, DepNeg produced higher F-score and accuracy than cTAKES Negation. Both models tend to perform better in precision than recall and this phenomenon is especially noticeable in DepNeg. In DepNeg recall is lower but precision is much higher than cTAKES Negation (0.8462 in cTAKES vs. 0.9665 in DepNeg in All) – i.e., DepNeg produced much lower number of FPs than cTAKES Negation. In signs/symptoms, which contain higher ratio of negated entities, DepNeg produced very high precision (0.9884) with the same level of recall in cTAKES negation.
Discussion
DepNeg produced better negation performance than the original cTAKES negation in terms of F-score and accuracy. This improvement is mainly due to reducing FPs and so increasing precision. Since the negation scope using dependency rules is not limited to by distance (i.e., number of word tokens) but depends on syntactic information in context, DepNeg was able to correctly determine more complicated negations than cTAKES negation.
DepNeg performed more accurately than the cTAKES negation in following cases where DepNeg determinations were correct but cTAKES negations were incorrect. A bold denotes a named entity to be examined for negation:
Contextual understanding (truth state = not negated): e.g., “He felt that no specific therapy was available regarding Moebius sequence.” “I do not recommend drug treatment for stone prevention.” “If her pain should not have been resolved by that time, there is the possibility of repeating facet rhizotomy.”
Long distance between named entities and negation words (truth state = negated): e.g., “She denies any ear pain, sore throat, odynophagia, hemoptysis, shortness-of-breath, dyspnea on exertion, chest discomfort, anorexia, nausea, weight-loss, mass, adenopathy or pain.”
Determining the direction of negation scope (truth state = not negated): e.g., “However, I suspect that her pain is not due to an underlying neurologic disorder.”
However, DepNeg may fail in certain cases. The following are error analyses for examples of DepNeg failing to correctly identify negation while cTAKES negation was correct (truth state = negated):
Incorrect dependency parse: e.g., “Molecular fragile-X results reveal no apparent PMR-1 gene abnormality.” (The dependency parser incorrectly identified “no” as modifying “PMR” instead of “abnormality.) “She denies any blood in the stool.” (The dependency parser inferred that “in the stool” was a modifier of “denies” rather than “blood.”)
Incomplete rules: e.g., “Mrs. Jane Doe returns with no complaints worrisome for recurrent or metastatic oropharynx cancer.” (The syntactic construction “worrisome for...” is not present as a rule in dependency path patterns.)
Both 1) and 2): e.g., “She is not having any incontinence or suggestion of infection at this time” (“Of infection” was parsed incorrectly to modify “incontinence” instead of “suggestion” The second Negated Noun rule (see Figure 4) should be generalized to allow conjunctions with constructions like “suggestion of,” where the focus is in a prepositional phrase.)
Dependency pattern rules were developed by examining a small subset of data from the 2010 I2B2/VA NLP challenge. We believe that more variety of dependency pattern rules derived from a larger data set can reduce type-2 errors. As can be seen in the type-1 errors), the performance of DepNeg is also affected by the accuracy of the dependency parser. The cTAKES dependency parser learns a dependency structure from training data and therefore different training sets might vary performance. Based on our observations, training dependency parsers on larger corpora produced higher negation detection accuracy in DepNeg. When using a dependency parser trained only on clinical questions instead of the merged sets of three data sources, DepNeg produced lower negation accuracy due to the inferior dependency parse performance. Although the training set of clinical questions might be in the same domain with respect to semantic content, its syntactic constructions were highly divergent. In this sense, we believe that using a dependency parser trained on more comprehensive data would reduce type-1 errors.
Conclusion
The main focus of this study was to examine the possibility of enhancement in negation annotation when using dependency structure rather than to maximize negation accuracy. Our results support the idea that dependency structure can improve the negation annotation that uses a fixed negation scope. Even though we have used a limited set of dependency rules compiled from a small data set, DepNeg performed well and was able to identify complicated negations that were wrong in the original cTAKES negation. In the future we believe that more comprehensive dependency rules that cover most negation patterns and a dependency parser trained on additional data would further improve negation accuracy. We also believe that using more negation words compiled from extra data or sources could enhance the DepNeg performance. Finally, as more negation-annotated clinical data becomes available, machine learning techniques may be used in conjunction with our heuristic rules defined here to further improve negation accuracy.
Acknowledgments
We would like to thank Vinod Kaggal who provided us with technical support to modifying cTAKES negation annotator. This work was partially supported by SHARP 4 grant 90TR000201 (PI Chute).
Footnotes
a corpus of 2000 clinical questions that have been treebanked according to the up-to-date Penn Treebank guidelines
References
- 1.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. Evaluation of negation phrases in narrative clinical reports. Proc AMIA Symp. 2001;2001:105–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Savova G, Masanz J, Ogren P, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savova GK, Kipper-Schuler K, Buntrock JD, Chute CG. UIMA-based clinical information extraction system. LREC 2008: Towards enhanced interoperability for large HLT systems:UIMA for NLP. 2008;2008 [Google Scholar]
- 4.D'Avolio LW, Nguyen TM, Farwell WR, et al. Evaluation of a generalizable approach to clinical information retrieval using the automated retrieval console (ARC) J Am Med Inform Assoc. 2010;17(4):375–82. doi: 10.1136/jamia.2009.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen RD, Masanz J, Ogren P, Ward W, Martin JH, Savova GK. An Architecture for Complex Clinical Question Answering (in press) IHI. 2010;2010 [Google Scholar]
- 7.Sohn S, Savova GK. Mayo Clinic Smoking Status Classification System: Extensions and Improvements. AMIA Annual Symposium; 2009; San Francisco, CA. 2009. pp. 619–623. [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 9.Mutalik P, Deshpande A, Nadkarni P. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc. 2001;8:598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkin PL, Brown SH, Bauer BA, et al. A controlled trial of automated classification of negation from clinical notes. BMC Medical Informatics and Decision Making. 2005;5(13) doi: 10.1186/1472-6947-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldin M, Chapman WW. Learning to detect negation with ‘Not’ in medical texts. Proceedings of ACMSIGIR. 2003;2003 [Google Scholar]
- 12.Huang Y, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assoc. 2007;14(3):304–11. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruch P, Baud R, Geissbuhler A, Rassinoux A. Comparing general and medical texts for information retrieval based on natural language processing: an inquiry into lexical disambiguation. Medinfo. 2001;10:261–5. [PubMed] [Google Scholar]
- 14.Choi JD, Nicolov N. Amsterdam & Philadelphia: John Benjamins; 2009. K-best, Locally Pruned, Transition-based Dependency Parsing Using Robust Risk Minimization. [Google Scholar]
- 15.Choi JD, Palmer M. Getting the most out of transition-based dependency parsing. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies (ACL:HLT'11); 2011; Portland, Oregon. 2011. pp. 687–92. [Google Scholar]
- 16.Nivre J, Hall J, Nilsson J. MaltParser: A Data-Driven Parser-Generator for Dependency Parsing. Proceedings of the fifth international conference on Language Resources and Evaluation (LREC2006); 2006; Genoa, Italy. 2006. pp. 2216–9. [Google Scholar]
- 17.Marneffe M-Cd, MacCartney B, Manning CD. Generating Typed Dependency Parses from Phrase Structure Parses. Proceedings of the fifth international conference on Language Resources and Evaluation (LREC2006); 2006; Genoa, Italy. 2006. pp. 449–54. [Google Scholar]
- 18.Tateisi Y, Yakushiji A, Ohta T, Tsujii Ji. Syntax Annotation for the GENIA corpus. Proceedings of the IJCNLP; 2005; Jeju Island, South Korea. 2005. pp. 222–7. [Google Scholar]
- 19.Marcus MP, Santorini B, Marcinkiewicz MA. Treebank-2. Philadelphia: Linguistic Data Consortium; 1995. [Google Scholar]