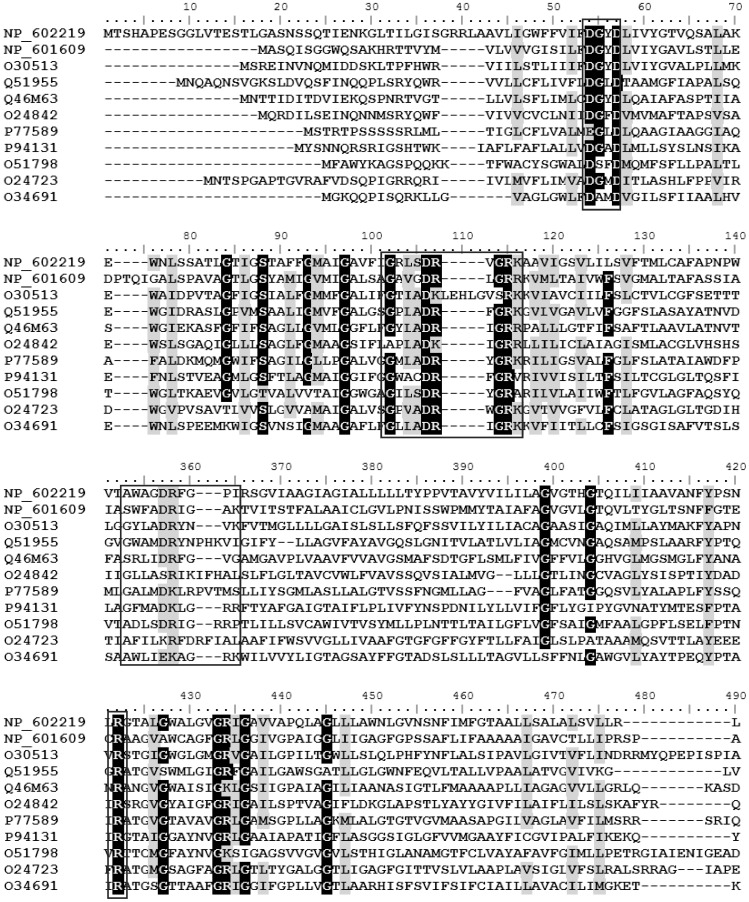
Figure 2. Sequence alignments of the AAHS family members by Clustal W.
The consensus profiles identified are highlighted. The black-marked residues indicate identical amino acid residues and the gray-marked residues indicate similar amino acid residues with a similarity threshold of 75%. The residues for site-directed mutagenesis are in the motifs which are surrounded in the rectangle. The accession numbers of all the AAHS family members in the TCDB database (http://www.tcdb.org/) for comparison are as follows: GenK (accession no. NP_602219), BenK (accession no. NP_602219 and O30513), PcaK (accession no. Q51955), TfdK (accession no. Q46M63), VanK (accession no. O24842), MhpT (accession no. P77589), MucK (accession no. P94131), MmlH (accession no. O51798), Orf1 (accession no. O24723), YceI (accession no. O34691).