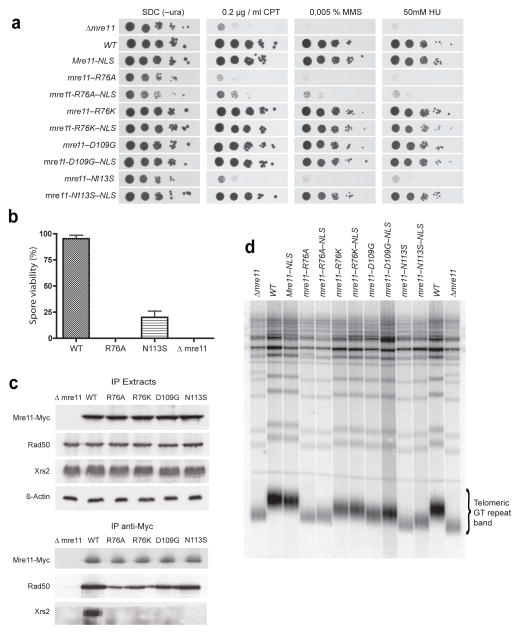
Figure 5. In vivo characterization of Mre11 latching loop compromising mutations in S. cerevisiae.
(a) Plate survival assays by serial dilutions reveal sensitivity of latching loop targeting mutations to camptothecin (CPT), methyl–methanesulfonate (MMS) and hydroxyurea (HU).
(b) Spore viability is strongly reduced in ScMre11 R76A and ScMre11 N113S diploid strains. Spore viability was calculated as the percentage of viable spores on total spores after tetrad dissection. Error bars represent s.e.m. spore viability obtained in two independent experiments, with 64 spores analyzed for each strain in each experiment.
(c) Mre11–Rad50–Xrs2 complex formation defects of Mre11 latching loop targeting mutations tested by co–immunoprecipitation. Cell extracts were immunoprecipitated with an anti–Myc antibody against Mre11-Myc and proteins were visualized by Western blotting with anti–Myc (Mre11), anti–Rad50, and anti–Xrs2 antibodies respectively. β–Actin was included as a loading control.
(d) Telomere lengths of Mre11 latching loop targeting mutations. Southern blot of XhoI–digested yeast DNA probed with a poly(GT)20 oligonucleotide specific for telomeric repeats is shown. The bracket indicates the telomeric GT repeat band derived from Y′ element–containing chromosomes.