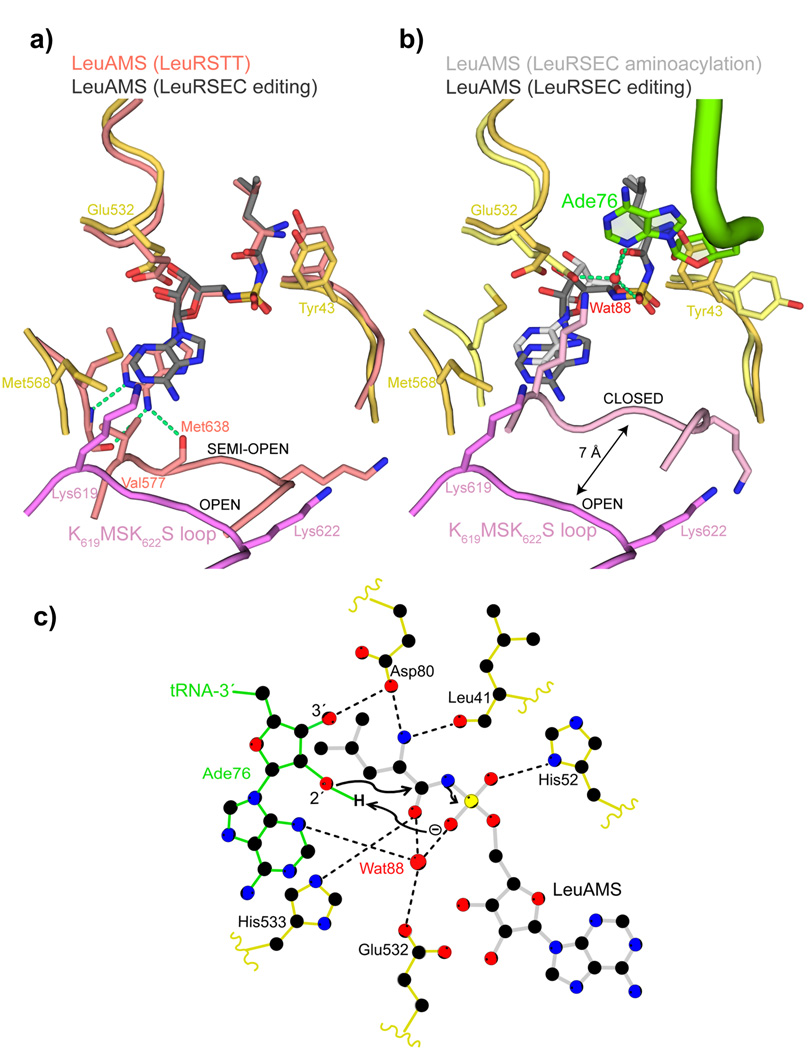
Figure 4. tRNA 3′ end binding induces full closure of the KMSKS loop.
a. Comparison between the synthetic sites of the LeuRSEC-LeuAMS-tRNALeu complex with the tRNA in the editing configuration (catalytic domain yellow, fully open KMSKS loop purple, LeuAMS dark grey) and the LeuRSTT-LeuAMS complex (PDB 1H3N) with the KMSKS loop in the semi-open conformation (all chains and LeuAMS salmon). Hydrogen bonds between the semi-open KMSKS loop conformation and the adenine base of the LeuAMS are indicated by green dotted lines but are absent in the open conformation.
b. Comparison between the synthetic sites of the LeuRS-LeuAMS-tRNALeu complex in the aminoacylation configuration (catalytic domain pale yellow, closed KMSKS loop light purple, LeuAMS light grey, tRNA green) and editing configuration (as in (a)). In the aminoacylation state, the presence of Ade76 necessitates a side-chain flip of Tyr43 and a water molecule (Wat88, red sphere) is coordinated by Ade76 N3, the re-orientated Glu532 carboxylate, a LeuAMS sulphate oxygen and the carbonyl-oxygen of the substrate leucine as indicated by green dotted lines (see text and (c)).
c. Schematic diagram of the LeuRSEC active site indicating the presumed substrate assisted mechanism of aminoacylation (see text).