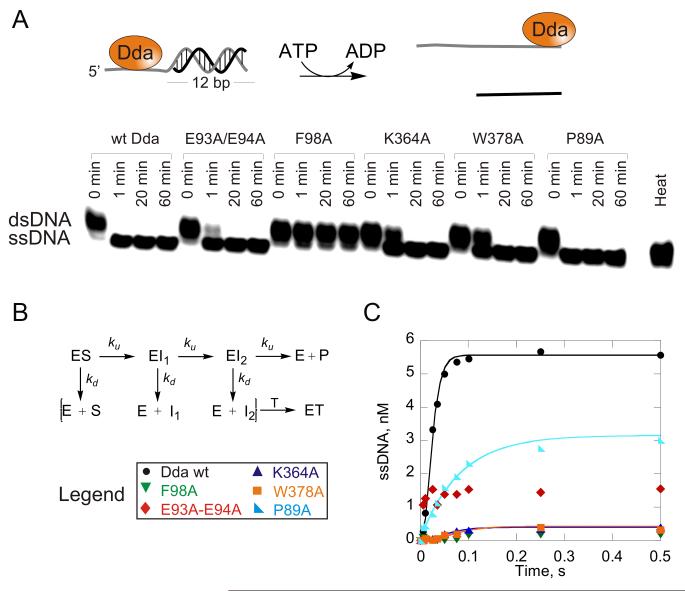
Figure 5. DNA unwinding activity.
(A) DNA Unwinding under multiple-cycle conditions. DNA, 10 nM dT8:12bp substrate was incubated with helicase (100 nM) and the reaction was initiated by mixing with ATP, Mg2+, phosphoenol pyruvate, pyruvate kinase/lactate dehydrogenase, and DNA trap. ssDNA was separated from dsDNA on a 20% native polyacrylamide gel and visualized by phosphorimager analysis. (B) A 16 bp DNA substrate is unwound by Dda in a three-step unwinding scheme which was used to fit data using KinTek Explorer (Johnson et al., 2009). Starting with enzyme (E) and substrate (S) prebound, the enzyme can convert the substrate to product in a series of three identical steps, defined by rate constant (ku). At each step, the enzyme can also dissociate from the substrate with rate constant kd and bind to the DNA trap (T). (C) DNA unwinding under single-cycle conditions. Unwinding of 10 nM dT7:16bp substrate by 1 μM Dda. Enzyme and substrate were pre-incubated, and the reaction was initiated by mixing with ATP, Mg2+, DNA trap, and protein trap. Data were fit to the three-step sequential mechanism shown in B for wt, W378A and K364A forms of Dda. A single exponential was used to fit unwinding data for the P89A variant. The rate constants and amplitudes of unwinding are listed in Table 2. See also Figure S2.