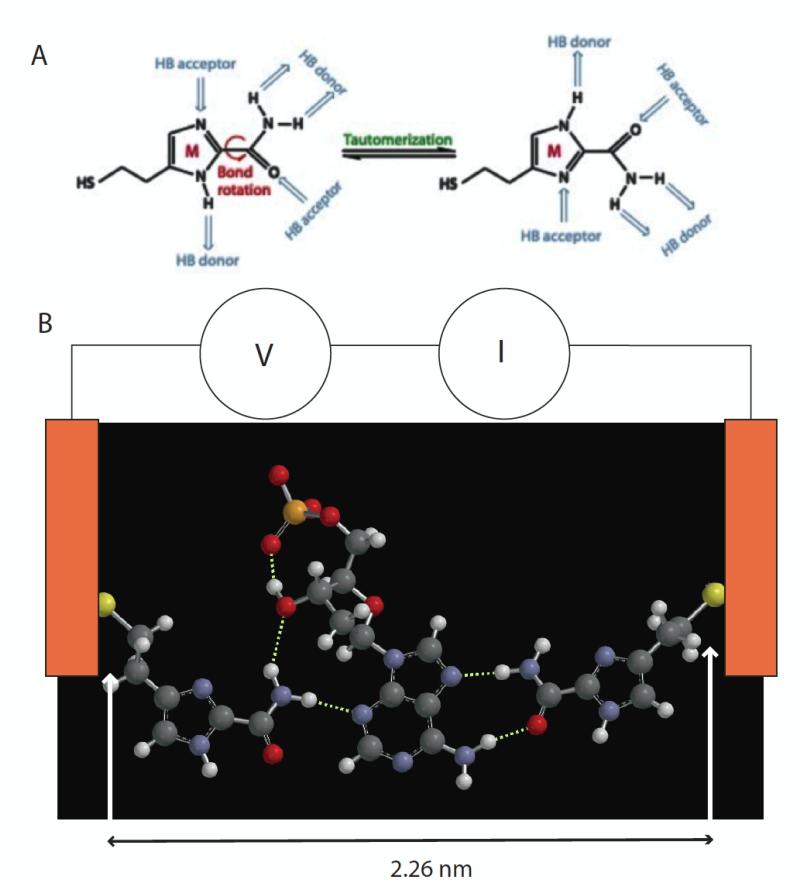
Figure 1.
(A) 4(5)-(2-mercaptoethyl)-1H-imidazole-2-carboxamide adaptor molecules (referred to as “M” in the text) showing how tautomerization presents different arrangements of hydrogen bond donors and acceptors. (B) This adaptor molecule (left and right side) trapping dAMP (middle) via a network of hydrogen bonds (dotted white lines). The sulfur atoms are bonded to gold electrodes, and current, I, is read as a bias, V, is applied across the tunnel gap. Individual 2D chemical structures were exported to Spartan’10 (Wavefunctions Inc.) to generate corresponding 3D structures that were energy minimized using the built-in MMFF molecular mechanics prior to the DFT calculation. All the structures were first calculated using B3LYP with /6-31G* in vacuum (structures for all four nucleotides are shown in Fig. S1). The gap size is set by the tunnel conductance and either maintained under servo control or left uncontrolled (but monitored via the baseline current).