Abstract
Objectives
To study ontology modularization techniques when applied to SNOMED CT in a scenario in which no previous corpus of information exists and to examine if frequency-based filtering using MEDLINE can reduce subset size without discarding relevant concepts.
Materials and Methods
Subsets were first extracted using four graph-traversal heuristics and one logic-based technique, and were subsequently filtered with frequency information from MEDLINE. Twenty manually coded discharge summaries from cardiology patients were used as signatures and test sets. The coverage, size, and precision of extracted subsets were measured.
Results
Graph-traversal heuristics provided high coverage (71–96% of terms in the test sets of discharge summaries) at the expense of subset size (17–51% of the size of SNOMED CT). Pre-computed subsets and logic-based techniques extracted small subsets (1%), but coverage was limited (24–55%). Filtering reduced the size of large subsets to 10% while still providing 80% coverage.
Discussion
Extracting subsets to annotate discharge summaries is challenging when no previous corpus exists. Ontology modularization provides valuable techniques, but the resulting modules grow as signatures spread across subhierarchies, yielding a very low precision.
Conclusion
Graph-traversal strategies and frequency data from an authoritative source can prune large biomedical ontologies and produce useful subsets that still exhibit acceptable coverage. However, a clinical corpus closer to the specific use case is preferred when available.
Keywords: Controlled, controlled terminologies and vocabularies, data bases, developing and refining EHR data standards (including image standards), knowledge bases, knowledge representations, measuring/improving patient safety and reducing medical errors, medical records, natural-language processing, ontology, other methods of information extraction, semantic web, SNOMED CT, software, systematized nomenclature of medicine, vocabulary
Due to their growing complexity and size, biomedical ontologies represent a serious challenge to creators, maintainers, and potential users. One of the most prominent examples of this type of ontology is SNOMED CT, the largest ontology project in the biomedical domain.1
SNOMED CT, which is intended to provide comprehensive, multilingual terminology for encoding all aspects of electronic health records, is based on a taxonomy of more than 390 000 concepts linked to terms and multilingual synonyms. In addition to this terminological component, SNOMED CT exhibits a language-independent ontological layer composed of a large polyhierarchic taxonomic backbone enriched by formal axioms that connects concepts across the hierarchies and supplies necessary and (partly) sufficient criteria formulated as description logic axioms conforming to the EL++ standard.2 3 Although SNOMED CT describes itself as a terminology, because it is expressed in a description logic and shows strong ontological commitment it is often referred of as an ontology.4
SNOMED CT provides a mechanism to build specific vocabularies defined by users, referred to as subsets (or reference sets in the new release format 2). Subsets have been suggested as being the key to making SNOMED CT usable,5 and their representation and management are thoroughly explained in SNOMED CT's technical documentation.6
The advisability of using only a part of rather than the whole ontology is not specific to SNOMED CT. In fact, the increasing size and complexity of ontologies has resulted in the establishment of an independent area of research known as ontology modularization, or decomposition.7 Reasons for modularization include computability (eg, scalability for querying data and reasoning, scalability for evolution and maintenance, and quality assurance) as well as usability (eg, understandability, context-awareness, personalization, and reuse).7
Ontology subsets are also referred to as segments, partitions, or modules. Given an input set of terms, known as seeds, target nodes, or signatures, modularization techniques identify terms that are related to the signature and that are therefore expected to be of interest for that particular use case. Graph-traversal modularization employs link-traversal heuristics to collect terms and axioms related to the signature, considering the ontology itself a graph;8–11 logic-based approaches use the underlying logic describing the ontology to collect axioms that guarantee safe reuse of the signature for reasoning purposes.12
Evaluating the performance or optimality of ontology modules has proved to be extremely difficult. Cuenca Grau et al12 acknowledged that their ‘experiments may not necessarily reflect an actual ontology reuse scenario’. After attempting to establish a set of criteria to determine the quality of a module, D'Aquin et al13 concluded that ‘there is no universal way to modularize an ontology’ and that ‘the choice of a particular technique or approach should be guided by the requirements of the application or scenario relying on modularization’.
However, modularization is particularly relevant to biomedical informatics, as biomedical ontologies are among the largest and most complex ontologies ever developed. Furthermore, due to the high degree of specialization in healthcare and life sciences, very few users require the whole breadth of these extended biomedical ontologies.14 By using modules in applications, such as annotation, an increase in accuracy, consistency and speed is expected. Therefore, modularization use cases related to semantic annotation are broad, ranging from clinical notes for intensive care services to medical images.15 16
However, neither the link-traversal nor logical methods may be satisfactory for biomedicine in general, and for annotation use cases in particular. An alternative approach is to extract a representative module (eg, a module providing high coverage when annotating) rather than a minimal module.12 For this reason, biomedical researchers have explored other approaches that do not rely on ontology structure or logic, but instead depend on related external information. For example, the CORE problem list subset for SNOMED CT is a subset of 5814 SNOMED CT concepts built with the help of seven health institutions worldwide. It contains the terms most frequently used when annotating clinical information at a summary level in several disciplines.17 Patrick et al15 developed a SNOMED CT subset after analyzing 44 million patient progress notes at an intensive care service. In this case, a specific logic-based tool to post-process the module was used.
Frequently, these restricted preconditions cannot be met: extensive case-specific pre-existing information might be scarce or even non-existent in many cases, and a specific tool might not be available, or could be tied to a particular ontology. Under these conditions, frequency-based filtering using an authoritative corpus can be used as an alternative to reduce the size of an ontology or a subset. Concepts that do not reach a certain threshold are considered irrelevant and can be filtered out. As an example, the united medical language system (UMLS) terminology has been filtered using information from MEDLINE with encouraging results.18
Through this study, we aimed to understand better what results are to be expected when graph-traversal and logic-based ontology modularization techniques are applied to a large biomedical terminology, such as SNOMED CT, under the specific conditions of an annotation scenario in which no previous corpus to be analyzed exists. We were also interested in evaluating the coverage-neutral reduction of the extracted modules' size by filtering them using an authoritative external corpus. To our knowledge, the approach and comparative results described in this report are the first of their kind.
Objectives
The main objectives of this study are:
(2) To explore whether a combination of (1.1) and (1.2) with (1.3) can extract better modules in terms of size, coverage, and precision.
As a use case for evaluation, we present an annotation scenario with the following aims: to create a SNOMED CT module, which we term a SNOMED CT M module (for a domain of discourse D), that is (a) significantly (ie, one order of magnitude) smaller, and (b) provides high coverage of D. Furthermore, (c) the fragment should preserve the logical entailments that can be derived from the original ontology as much as possible.
However, for the current use cases (c) is a secondary goal, as SNOMED CT's routine use has thus far been restricted to the provision of controlled terms, given the preliminary and still controversial status of many axioms and the structure of the hierarchies.21–23
Materials and methods
We have chosen annotation of cardiology discharge summaries as our domain of discourse. These summaries contain information such as the reasons for admission, past history, interventions, and proposed follow-up.
When compared with classic modularization approaches, the task here is not to extract a minimal module, but to extract a representative module.12 We expected different resulting SNOMED CT M modules for the same typical, but not exhaustive, input signatures for D. For instance, the signatures may include several SNOMED CT concepts that represent typical cardiovascular drugs, and M is predicted to include additional drugs that are likely to be prescribed for cardiovascular disorders, but not, for example, chemotherapeutic agents used in the treatment of cancer.
Experimental design
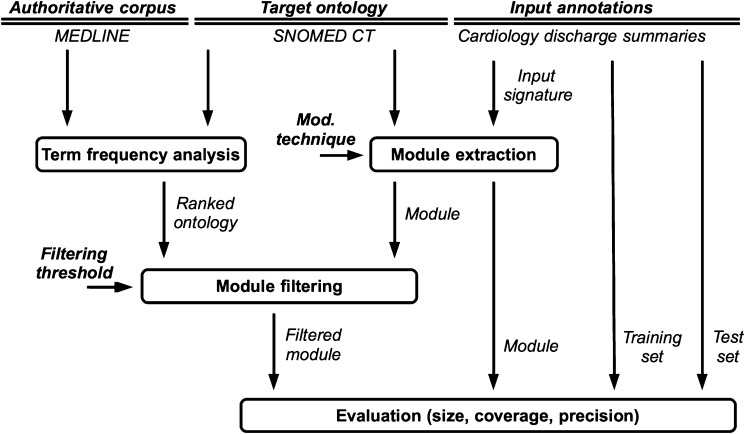
Figure 1 shows the experimental arrangement of (1) the inputs (the SNOMED CT ontology to be modularized, the reference authoritative corpus used to filter the modules, and the input annotations to provide signatures and to measure coverage); (2) the processes (module extraction, term frequency analysis and filtering, and evaluation); (3) the parameters (modularization technique and filtering threshold); and (4) the measured outputs (module size, coverage, and precision). The rest of this section describes the experimental processes in detail.
Figure 1.
Experimental set-up, design, parameters and measured variables.
Module extraction using ontology modularization techniques
The January 2010 international release of SNOMED CT was used as the target ontology for modularization. This release contained 390 022 concepts; of these, 291 205 were marked as active, and the remaining concepts were retained for backwards compatibility.
To extract subsets using graph-traversal heuristics, we followed the approach proposed by Seidenberg and Rector.9 Their method, ontology segmentation, employs a heuristic to traverse a graph for the purpose of obtaining a subset, or segment, of related nodes, starting from one or more input nodes of interest, which are referred to as seeds, target nodes or signatures.
Signatures for the experiments were obtained from a corpus of 20 discharge summaries. This corpus was written in Portuguese and authored by physicians of the Hospital de Clínicas de Porto Alegre, which is a large university hospital in Porto Alegre, Brazil. In the context of an information extraction project, all medical terms appearing in each summary had been manually coded using the concept identifiers of the most fine-grained terms in SNOMED CT. The text appearing in each summary contained reasons for admission, past history, interventions, and the proposed follow-up of cardiology inpatients. The number of SNOMED CT concepts required to annotate the discharge summaries ranged from 17 to 64, with an average of 35. A total of 439 different SNOMED CT concepts was needed to annotate the complete set of 20 summaries. A sample fragment of an annotated discharge summary is shown in table 1.
Table 1.
Sample fragment of an annotated discharge summary
| Original text | Most fine-grained SNOMED CT term | SNOMED CT concept ID |
| Masculino | Male (finding) | 248153007 |
| 43 anos | Current chronological age (observable entity) | 42414402 |
| Hipertenso | Hypertensive disorder, systemic arterial (disorder) | 38341003 |
| Tabagista | Tobacco user (finding) | 110483000 |
| Etilista | Current drinker of alcohol (finding) | 219006 |
| Interna | Hospital admission (procedure) | 32485007 |
| Por infarto agudo do miocárdio | Acute myocardial infarction (disorder) | 57054005 |
| Sem supradesnivelamento de segmento ST | ST segment elevation (finding) | 76388001 |
The terms appearing in each summary had been manually coded using the concept identifiers of the most fine-grained terms in SNOMED CT.
The discharge summaries were also used to evaluate the estimated coverage of the extracted subsets. Complete information regarding the use of the annotations can be found in the Evaluation section.
The heuristic proposed by Seidenberg and Rector9 first builds a set containing the complete hierarchy of the target node and then recursively follows links for every node in the set. However, depending on the size and topology of the ontology and target nodes, this method might not be useful in practice for both performance and size reasons: Seidenberg and Rector9 had to introduce two strategies, property filtering and depth limiting, to limit the segment size. Furthermore, Doran et al10 proposed a modification of the heuristic that prevented upwards navigation of the taxonomy from the target node, based on the justification that this would increase the probability of extracting modules as large as the whole ontology.
Taking these strategies into account, we developed four graph-traversal heuristics of increasing complexity to collect concepts of interest but limit the size of the extracted subset.
Upwards segmentation: A modification of the heuristic described by Seidenberg and Rector,9 where is-a and attribute links from the concepts in the signature are followed upwards in the hierarchy, with the root on top. However, our variation does not recursively repeat the process for every node in the sub-trees of concepts in the signature.
S-heuristic: This heuristic follows the same strategy as the upwards segmentation but adds the sibling nodes of signature concepts.
ST-heuristic: This is the same as the S-heuristic, also including the complete sub-trees of all added siblings.
IL-heuristic: Similar to the ST-heuristic, it adds all nodes that are connected to the signature concepts using linkage concepts.
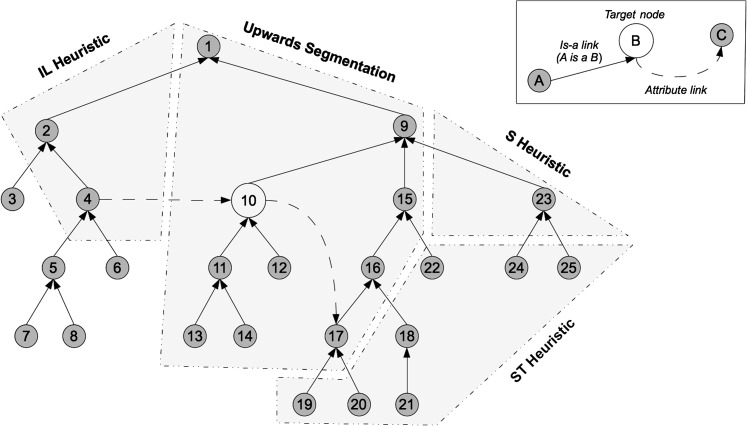
An example of the nodes collected by each heuristic with respect to the previous one is shown in figure 2. The heuristics are incremental: nodes collected by the upwards segmentation heuristic are also collected by the S-heuristic, while nodes collected by the S-heuristic are also collected by the ST-heuristic, and so on. Further details about the heuristics can be found in supplementary appendix A (available online only).
Figure 2.
Example ontology showing nodes identified by each of the graph-traversal heuristics.
To evaluate the locality logic-based modularization technique of Cuenca Grau et al,12 we used the tool provided by the authors.24 The intuitive idea behind locality is to identify and discard all axioms from an ontology that are logically irrelevant to the input signature. The resulting module can then be safely used as a substitute of the whole ontology when referring to symbols from the signature (see supplementary appendix B, available online only, for details). Locality-based modularization is now part of the ontology web language application programming interface (OWL API).
Term frequency analysis and module filtering
The aim of the filtering process is to identify relevant nodes according to MEDLINE. Given an input set of concepts, the filtering process selects or discards each input concept depending on its precomputed score in the ranked ontology and the threshold set by the user. A threshold of one selects all concepts that appear at least once in the local MEDLINE repository; of two, concepts that appear twice, etc. A threshold of zero indicates no filtering.
We built a ranked version of SNOMED CT, following the approach of Xu et al.18 Although other sources of synonyms exist (eg, the UMLS metathesaurus), the scope of our project was limited to SNOMED CT; thus, SNOMED CT terms were obtained from the descriptions file included in SNOMED CT as distributed by International Health Terminology Standards Development Organisation (IHTSDO). The file contains all terms linked to SNOMED CT concepts, ie, preferred terms and (quasi-)synonyms. As an example, 11 terms are accepted for concept identifier 22298006 (‘myocardial infarction’), including ‘heart attack’, ‘cardiac infarction’ and ‘infarction of heart’. The SNOMED CT version used in this study contained 1 157 834 descriptions for 390 022 concepts.
For each SNOMED CT concept, we counted the number of occurrences of each term associated with the concept in either the title or abstract of a subset of MEDLINE articles (see the Term frequency analysis and module filtering in the Results section for details). Terms that did not reach a minimum input threshold were considered irrelevant to scientific discourse and therefore filtered out. Details on the complete process of term matching, SNOMED CT ranking and module filtering can be found in supplementary appendix C (available online only).
Evaluation
The effects of every modularization technique and filtering threshold on size, coverage, and precision were measured. The detailed configuration of the 10-fold cross-validation experiments that were performed can be found in supplementary appendix D (available online only).
As values for the modularization parameter, we used the five techniques presented in the Materials and methods section: upwards segmentation, S-heuristic, ST-heuristic, IL-heuristic, and locality (LUM). The full SNOMED CT terminology and the CORE problem list subset were also added as useful references. Note that the CORE problem list subset was used as distributed, without any post-processing. As values for the filtering threshold parameter, we used 0, 1, 10, 100, 1000, 10 000, and 100 000.
The following variables were measured:
Size: Total number of concepts in the extracted subset in relation to SNOMED CT (390 022).
Coverage (or ‘recall’): Ratio of relevant concepts in a subset to the total number of (unique) concepts in the test set of patient summaries. A concept in the subset was considered relevant if its code occurred at least once in the test set.
Precision: Ratio of relevant concepts to the total number of concepts in the subset.
Results
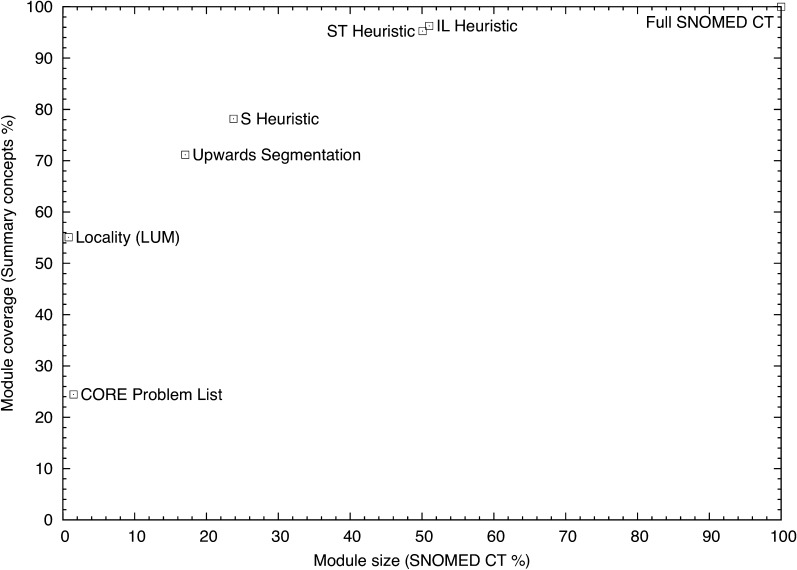
The average performance of the extracted subsets in terms of coverage, size, and precision over the results of the different test sets in the 10-fold cross-validation is shown in table 2 and figure 3. The full SNOMED CT dataset and the CORE problem list subset were added as references for comparison.
Table 2.
Average subset size, coverage and precision for the analyzed modularization techniques, in addition to the CORE problem list subset and the full SNOMED CT, which are included as references
| Modularization technique | Subset size (SNOMED CT %) | Subset coverage | Subset precision |
| CORE problem list | 1.49% | 24.44% | 0.27% |
| Locality (LUM) | 0.80% | 55.08% | 1.14% |
| Upwards segmentation | 17.40% | 71.15% | 0.07% |
| S-heuristic | 23.76% | 78.16% | 0.05% |
| ST-heuristic | 50.07% | 95.26% | 0.03% |
| IL-heuristic | 51.01% | 96.26% | 0.03% |
| Full SNOMED CT | 100.00% | 100.00% | 0.02% |
Figure 3.
Subset extraction using ontology modularization techniques.
Using graph-traversal techniques, the average coverage ranged from 71% to 96%, while the average module size ranged from 17% to 51%. The locality-based technique extracted the smallest (1%) and most precise (1.14%) module, but coverage was strongly affected (55%).
As can be seen in table 2, an increase from 1% (locality) to 23% (S-heuristic) in the module size raised the coverage to 78%. Doubling the module size to a total of 50% (ST-heuristic) added only another 20%, bringing the coverage up to 95%. Doubling the module size again to 100% using the full SNOMED CT only added the final 5% coverage or less. Therefore, 23% and 50% are two important sizes to consider depending on the coverage needs of the application (78% or 95%). Precision was extremely low in all cases (approximately 1% or lower). In order to preserve logical entailments, many auxiliary concepts must be included (eg, anatomical structure, organisms, values for qualifiers, etc). These auxiliary concepts never appear on their own in the discharge summaries, even though they typically outnumber the concepts in the original signature.
Term frequency analysis and module filtering
The first step to filter the previous subsets was to build a ranked version of SNOMED CT by analyzing the frequency of appearance of each of its terms in MEDLINE.
Through accessing the PubMed search engine, 206 484 records of interest were retrieved by searching for a subset containing human case reports written in English from the past 5 years. After parsing and insertion into Lucene, the indexing engine reported a total of 947 285 stored terms, with 120 657 appearing in the title field and 826 628 being found in the abstract.
After analyzing and matching each active SNOMED CT term, a total of 43 550 different concepts was found to have been cited in the MEDLINE subset. The frequency of each matched term was added to the ranked copy of SNOMED CT, and the frequency data were then used to filter the subsets with thresholds of 1, 10, 100, 1000, 10 000, and 100 000.
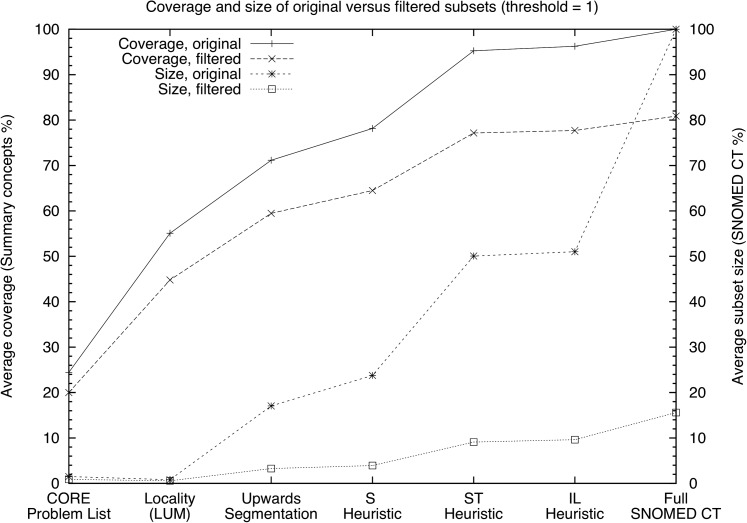
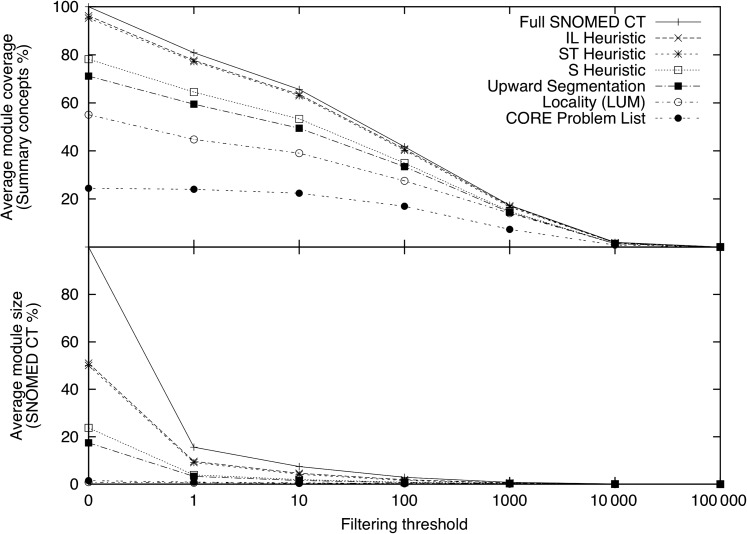
Figure 4 compares the performance of the original subsets and a filtered copy using a threshold of one. After filtering, the IL and ST-heuristics both showed a dramatic reduction in module size (from 50% to 9%) while still providing more than 77% coverage. When filtering the whole SNOMED CT, a module with a size of 15% providing coverage of 81% was obtained.
Figure 4.
Comparison of unfiltered versus filtered modules with a threshold of 1.
Table 3 shows the influence of filtering thresholds ranging from 0 (no filtering) to 100 000 on subset size, coverage, and precision. Filtering improved precision in all cases, and a threshold of 1000 provided the highest value for this parameter (except for the CORE problem list, in which 10 000 provided higher precision).
Table 3.
Influence of filtering thresholds on subset size, coverage, and precision
| Filtering threshold | 0 | 1 | 10 | 100 | 1000 | 10 000 | 100 000 |
| CORE problem list | |||||||
| Module size | 1.49% | 0.88% | 0.58% | 0.22% | 0.03% | <0.01% | <0.01% |
| Coverage | 24.44% | 24.00% | 22.41% | 16.95% | 7.36% | 0.80% | <0.01% |
| Precision | 0.27% | 0.45% | 0.64% | 1.27% | 3.64% | 8.33% | <0.01% |
| Locality (LUM) | |||||||
| Module size | 0.80% | 0.57% | 0.45% | 0.31% | 0.15% | 0.02% | <0.01% |
| Coverage | 55.08% | 44.80% | 39.07% | 27.53% | 14.07% | 1.40% | <0.01% |
| Precision | 1.14% | 1.29% | 1.41% | 1.45% | 1.51% | 1.01% | <0.01% |
| Upwards segmentation | |||||||
| Module size | 17.40% | 3.26% | 1.54% | 0.64% | 0.24% | 0.04% | <0.01% |
| Coverage | 71.15% | 59.47% | 49.45% | 33.44% | 14.45% | 1.40% | <0.01% |
| Precision | 0.07% | 0.30% | 0.52% | 0.85% | 0.98% | 0.59% | <0.01% |
| S-heuristic | |||||||
| Module size | 23.76% | 3.93% | 1.94% | 0.84% | 0.31% | 0.05% | <0.01% |
| Coverage | 78.16% | 64.50% | 53.30% | 34.99% | 14.85% | 1.40% | <0.01% |
| Precision | 0.05% | 0.27% | 0.45% | 0.68% | 0.78% | 0.44% | <0.01% |
| ST-heuristic | |||||||
| Module size | 50.07% | 9.10% | 4.15% | 1.54% | 0.42% | 0.07% | <0.01% |
| Coverage | 95.26% | 77.17% | 62.97% | 40.26% | 16.64% | 1.76% | <0.01% |
| Precision | 0.03% | 0.14% | 0.25% | 0.43% | 0.64% | 0.42% | <0.01% |
| IL-heuristic | |||||||
| Module size | 51.01% | 9.62% | 4.60% | 1.86% | 0.51% | 0.07% | <0.01% |
| Coverage | 96.26% | 77.70% | 63.50% | 40.79% | 17.08% | 1.76% | <0.01% |
| Precision | 0.03% | 0.13% | 0.23% | 0.36% | 0.54% | 0.38% | <0.01% |
| Full SNOMED CT | |||||||
| Module size | 100% | 15.61% | 7.42% | 2.89% | 0.78% | 0.13% | 0.01% |
| Coverage | 100% | 80.87% | 65.51% | 41.73% | 17.38% | 1.94% | <0.01% |
| Precision | 0.02% | 0.08% | 0.14% | 0.24% | 0.36% | 0.23% | <0.01% |
A threshold of 0 indicates no filtering.
Figure 5 graphically displays the data presented in table 3. A dramatic reduction in subset size can be observed, even with the lowest filtering threshold, while there was only a modest loss of coverage.
Figure 5.
Influence of filtering in module coverage and size. A threshold of 0 indicates no filtering.
Discussion
Our case study revealed that a common scenario in biomedicine, the annotation of discharge summaries, can be particularly challenging for ontology modularization techniques that do not rely on additional information to improve accuracy or reduce subset size, such as using an external corpus for filtering. The main difficulty of the case study presented here is that the required subset, although it is compact and specific to a domain (cardiology), is still complex and is spread across a wide range of subhierarchies, as discharge summaries include information such as reasons for admission, past history, interventions, and proposed follow-up. When extracting modules, link-traversal techniques identify concepts from hierarchies (eg, intermediate concepts and sub-trees of target nodes, see supplementary appendix A, available online only), while logic-based techniques identify concepts that are parts of definitions or restrictions (see supplementary appendix B, available online only). Therefore, a number of concepts that might not be relevant for coding are nevertheless necessarily included in the subsets, severely affecting precision.
Several issues should be considered when comparing the performance of the CORE subset with the rest of our results: (1) the CORE subset is derived from problem lists that cover all of the major specialties in medicine, while the ontology modularization techniques presented here used concepts from a specific corpus (cardiology) as signatures; (2) in addition to findings, diagnoses and procedures, our test datasets also include drugs, laboratory tests and non-clinical concepts, none of which are within the scope of the CORE subset; and (3) the CORE subset cannot properly be considered a logically consistent module because it preserves neither the structure nor the logic of SNOMED CT. Post-processing to convert the CORE problem list into a module was performed by Rector et al,25 increasing the number of concepts from 8500 to 35 000, which is approximately 10% of the size of SNOMED CT. The public availability of this module in the SNOMED CT distribution format, along with the currently distributed plain list of problems, would be extremely useful to researchers in the field.
The number of SNOMED CT concepts found by Patrick et al15 in their corpus (30 000) is in good agreement with our observations (43 000). The results of Patrick et al15 (1% size, 96% coverage) suggest that when a large corpus specific to the use case exists, it should definitely be used as the primary source of information, in contrast to an external authoritative corpus. Unfortunately, a locally derived corpus is often not available.
Filtering was also employed by Xu et al18 to reduce the size of UMLS down to 13% using MEDLINE abstracts. When we employ filtering techniques and MEDLINE titles and abstracts to prune the full SNOMED CT, our results are similar (15% on average). However, filtering techniques require a preprocessing (matching, ranking, indexing) and post-processing workload (ontology reconstruction) that should be taken into account, as should the possible bias of the corpus. In our case, the most plausible explanation for the coverage of the filtered version of SNOMED CT (81%) is the content mismatch between clinical texts and scientific abstracts as well as the vocabulary mismatch between clinical jargon and scientific language. Nevertheless, the use of frequency data appears to be promising, and we will use a clinical corpus in the future.
When analyzing ontology modularization approaches, graph-traversal heuristics only require a small signature of concepts of interest and have been shown to provide decent coverage (from 71% to 96%), although at the expense of considerable subset size in some cases (from 17% to 51%). As expected when adding intermediate concepts into the hierarchy to preserve the entailments derived from the original ontology, precision is very low (<1%). However, this study provides evidence that when combined with the use of information on data frequency from a publicly available, authoritative source for filtering, graph-traversal techniques can prune large biomedical ontologies and produce subsets with an acceptable coverage. In our case, only 20 discharge summaries, but no pre-existing corpus, were used to provide the signatures for the ontology modularization techniques as well as to perform the evaluation. The limited number of discharge summaries employed should be taken into account when analyzing the results.
Although outside of the scope of our study, an open question of interest is whether the modules extracted by graph-traversal heuristics fulfil the safety requirements postulated by Cuenca Grau et al,12 ie, whether they produce exactly the same entailments as the complete SNOMED CT. All of the heuristics presented in this study preserve is-a relationships as well as cross-references from the concepts in the signature, and SNOMED CT axioms are rather uniform due to EL expressiveness and limited nesting. However, further investigation is needed if the subsets are to be used for reasoning purposes.
Conclusions and further work
A combination of graph-traversal strategies and information on data frequency from an external authoritative corpus can prune large biomedical ontologies and produce convenient subsets with fair coverage, without requiring a pre-existing corpus of information closely related to the use case or employing natural language processing techniques. However, how acceptable this coverage is depends on each specific use case.
In our future work, we will explore the following optimization strategies:
Size/coverage analysis differentiated by SNOMED CT sub-hierarchies, for example, findings, procedures, and substances;
Identifying the sections of SNOMED CT that could appear in a discharge summary from those that could not on their own (eg, ‘organism’, ‘anatomy’, etc);
Frequency data from real patient records to avoid terminology mismatches between the language of clinicians and researchers;
Use of novel ontology segmentation techniques currently being developed by the ontology modularization community.
Furthermore, different SNOMED CT coding scenarios may be distinguished. For instance, a scenario that allows concept post-coordination would probably require fewer concepts for a given coverage compared with the hitherto standard approach of only using pre-coordinated terms.
Extension to additional clinical disciplines would also be desirable. However, this would require the use of coded data covering the entire clinical process, which are still rare, or the use of natural language processing techniques for the automated annotation of medical summaries.
Problems when using precision in the classic sense suggest that improved metrics for utility and representativeness should be defined in future studies.
Finally, this study shows that the requirements of preserving entailments and achieving high precision necessarily conflict. The function of extracting entailment preserving modules, and of extracting most precise modules for use in coding patient data is different, and attempting to satisfy the two aims simultaneously can lead to unsatisfactory results.
Acknowledgments
The authors want to thank Ernesto Jiménez Ruiz and Bernardo Cuenca Grau for their contributions and help regarding logic-based modularization approaches; Mark Musen, and all the staff from the Stanford Center for Biomedical Informatics Research for their useful advice, reviews and feedback; the JAMIA reviewers for their constructive and helpful comments. They also thank the Hospital de Clínicas de Porto Alegre (Brazil) for the provision of a sample of discharge summaries, and the Catholic University of Paraná in Curitiba (Brazil) for the SNOMED CT annotations of selected summaries.
Footnotes
Funding: This work was supported by grant TIN2010-21387-C02-01 from the Ministerio de Ciencia e Innovación of the Spanish government.
Competing interests: None.
Contributors: PLG acquired the data, developed and adapted the algorithms, performed the analysis, and drafted the article. AI contributed to the discussion of the design of the study, supervised the experiments, and revised and enhanced the manuscript. SS and MB contributed to the conception and design of the study, supervised the experiments, and revised and enhanced the manuscript. All authors approved the final version to be published.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The 20 SNOMED CT coded discharge summaries used in this study are available for review upon request.
References
- 1.IHTSDO International Health Terminology Standards Development Organization. http://www.ihtsdo.org/ (accessed 20 Sep 2011).
- 2.Baader F, Calvanese D, McGuiness DL, et al. The Description Logic Handbook. Cambridge, UK: Cambridge University Press, 2003 [Google Scholar]
- 3.Baader F, Brandt S, Lutz C. Pushing the EL envelope. Nineteenth International Joint Conference on Artificial Intelligence; 30 July-5 August 2005, Edinburgh, Scotland, 2005:369 [Google Scholar]
- 4.Schulz S, Cornet R. SNOMED CT's Ontological Commitment. London, UK: Nature Publishing Group, Nature Precedings, 2009 [Google Scholar]
- 5.Benson T. Principles of Health Interoperability HL7 and SNOMED. Berlin, Germany: Springer-Verlag, 2010 [Google Scholar]
- 6.SNOMED CT Technical Implementation Guide. IHTSDO: The International Health Terminology Standards Development Organisation, http://www.ihtsdo.org/fileadmin/user_upload/doc/download/doc_TechnicalImplementationGuide_Current-en-US_INT_20110731.pdf (accessed 15 Nov 2011). [Google Scholar]
- 7.Stuckenschmidt H, Parent C, Spaccapietra S, eds. Modular Ontologies: Concepts, Theories and Techniques for Knowledge Modularization. Berlin, Germany: Springer-Verlag, 2009 [Google Scholar]
- 8.Noy NF, Musen MA. Specifying ontology views by traversal. Third International Semantic Web Conference; 7–11 November 2004. Hiroshima, Japan, 2004:713–25 [Google Scholar]
- 9.Seidenberg J, Rector A. Web ontology segmentation: analysis, classification and use. 15th International Conference on World Wide Web; 23–26 May 2006. Edinburgh, Scotland, UK, 2006:13–22 [Google Scholar]
- 10.Doran P, Tamma V, Iannone L. Ontology module extraction for ontology reuse: an ontology engineering perspective. 16th ACM Conference on Information and Knowledge Management; 6–10 November 2007. Lisbon, Portugal, 2007:13–22 [Google Scholar]
- 11.D'Aquin M, Schlicht A, Stuckenschmidt H, et al. Ontology modularization for knowledge selection. 18th International Conference on Information and Knowledge Management; 3–7 September 2007. Regensburg, Germany, 2007:874–83 [Google Scholar]
- 12.Cuenca Grau B, Horrocks I, Kazakov Y. Modular reuse of ontologies: theory and practice. J Artif Intell Res 2008;31:273–318 [Google Scholar]
- 13.D'Aquin M, Schlicht A, Stuckenschmidt H, et al. Criteria and evaluation for ontology modularization techniques. In: Stuckenschmidt H, Parent C, Spaccapietra S, eds. Modular Ontologies: Concepts, Theories and Techniques for Knowledge Modularization. Berlin, Germany: Springer-Verlag, 2009:67–89 [Google Scholar]
- 14.Pathak J, Johnson TM, Chute CG. Survey of modular ontology techniques and their applications in the biomedical domain. Integr Comput Aided Eng 2009;16:225–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patrick J, Wang Y, Budd P, et al. Developing SNOMED CT subsets from clinical notes for intensive care service. Health Care Informat Rev Online 2008;12:25–30 http://www.hinz.org.nz/journal/997 (accessed 20 Sep 2011). [Google Scholar]
- 16.Wennerberg P, Schulz K, Buitelaar P. Ontology modularization to improve semantic medical image annotation. J Biomed Inform 2011;44:155–62 [DOI] [PubMed] [Google Scholar]
- 17.Fung KW, McDonald C, Srinivasan S. The UMLS–CORE Project: a study of the problem list terminologies used in healthcare institutions. J Am Med Inform Assoc 2010;17:675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu R, Musen MA, Shah NH. A comprehensive analysis of five million UMLS metathesaurus terms using eighteen million MEDLINE citations. AMIA Annu Symp Proc 2010;2010:907–11 [PMC free article] [PubMed] [Google Scholar]
- 19.Cuenca Grau B, Horrocks I, Kazakov Y. Just the right amount: extracting modules from ontologies. 16th International Conference on World Wide Web; 8–12 May 2007. Banff, Alberta, Canada, 2007:717–26 [Google Scholar]
- 20.Cuenca Grau B, Halaschek-Wiener C, Kazakov Y. History matters: incremental ontology reasoning using modules. 6th International Semantic Web Conference; 11–15 November 2007. Busan, Korea, 2007:183–96 [Google Scholar]
- 21.Schulz S, Suntisrivaraporn B, Baader F, et al. SNOMED reaching its adolescence: ontologists' and logicians' health check. Int J Med Inform 2009;78(Suppl 1):S86–94 [DOI] [PubMed] [Google Scholar]
- 22.Schulz S, Cornet R, Spackman K. Consolidating SNOMED CT's ontological commitment. Appl Ontol 2011;6:1–11 [Google Scholar]
- 23.Rector AL, Brandt S, Schneider T. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. J Am Med Inform Assoc 2011;18:432–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiménez-Ruiz E, Cuenca Grau B, Sattler U, et al. Safe and economic re-use of ontologies: a logic-based methodology and tool support. 5th European Semantic Web Conference; 1–5 June 2008. Tenerife, Canary Islands, Spain, 2008:185–99 [Google Scholar]
- 25.Rector A, Iannone L, Stevens R. Quality assurance of the content of a large DL-based terminology using mixed lexical and semantic criteria: experience with SNOMED CT. 6th International Conference on Knowledge Capture; 26–29 June 2011. Banff, Alberta, Canada, 2011:57–64 [Google Scholar]