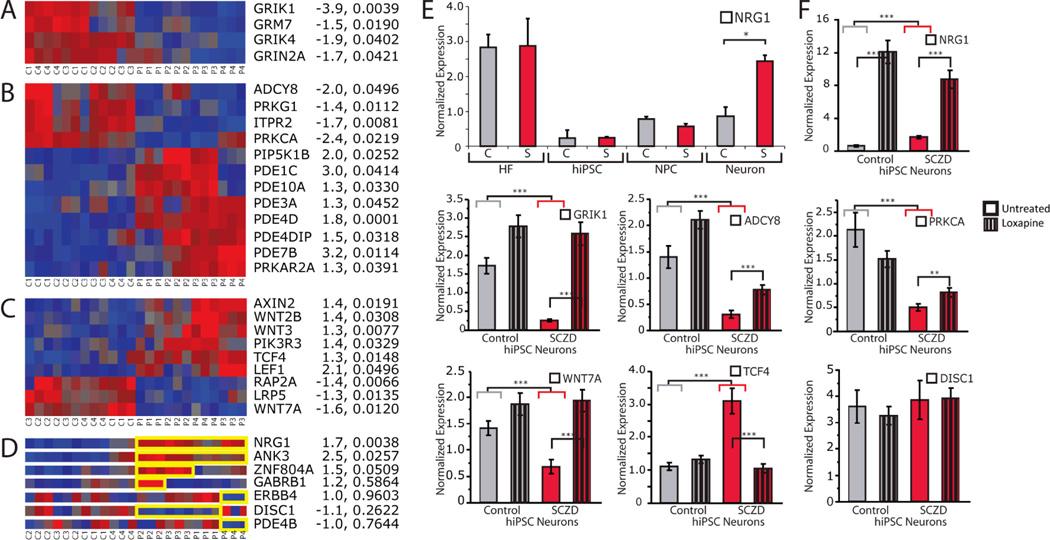
Fig. 4. RNA expression analysis of control and SCZD hiPSC neurons.
A–C. Heat maps showing microarray expression profiles of altered expression of glutamate receptors (A), cAMP signaling (B), and WNT signaling (C) genes in SCZD hiPSC neurons. Fold-change and p-values (diagnosis) provided to the right of each heat map. D. Heat maps showing perturbed expression (highlighted in yellow) of NRG1 and ANK3 in all four SCZD patients, as well as altered expression of ZNF804A, GABRB1, ERBB4, DISC1 and PDE4B in some but not all patients. Fold-change and p-values (diagnosis) provided to the right of each heat map. E. Altered expression of NRG1 is detected in SCZD hiPSC neurons but not in patient fibroblasts, hiPSCs or hiPSC NPCs. F. qPCR validation of altered expression of NRG1, GRIK1, ADCY8, PRKCA, WNT7A, TCF4 and DISC1, as well as response to three weeks of treatment with Loxapine (striped bars) in six-week-old hiPSC neurons. Asterisks used as follows: * p<0.05, ** p<0.01, *** p<0.001.