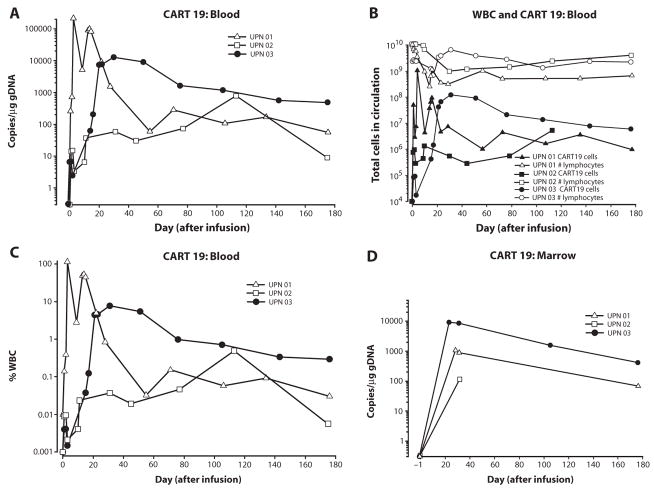
Fig. 2.
Sustained in vivo expansion and persistence in blood and marrow of CART19 cells. (A to D) qPCR analysis was performed on DNA isolated from whole blood (A to C) or bone marrow (BM) (D) samples obtained from UPN 01, UPN 02, and UPN 03 to detect and quantify CAR19 sequences. The frequency of CART19 cells is shown as average transgene copies (A), total calculated CART19 cells in circulation (B), or as a fraction of circulating white blood cells (WBCs) (C). (A) Copies CAR19/microgram DNA is calculated as described in Materials and Methods. (B) The total number of lymphocytes (total normal and CLL cells) versus total CART19+ cells in circulation is plotted for all three subjects using the absolute lymphocyte count from complete blood count values and assuming a 5.0-liter volume of peripheral blood. (C) % WBC is calculated as described in Materials and Methods. (D) Bulk qPCR analysis of marrow to quantify CART19 sequences. The data from patient UPN 03 in (A, C, and D) has been published in (9) and is reprinted here with permission. Each data point represents the average of triplicate measurements on 100 to 200 ng of genomic DNA, with maximal percent coefficient of variation (CV) less than 1.56%. Pass/fail parameters for the assay included preestablished ranges for slope and efficiency of amplification, and amplification of a reference sample. The lower limit of quantification for the assay established by the standard curve range was two copies of transgene per microgram of genomic DNA; sample values below that number are considered estimates and presented if at least two of three replicates generated a Ct value with percent CV for the values 15%. CART19 cells were infused at days 0, 1, and 2 for UPN 01 and 03 and at days 0, 1, 2, and 11 for UPN 02.