Fig 4.
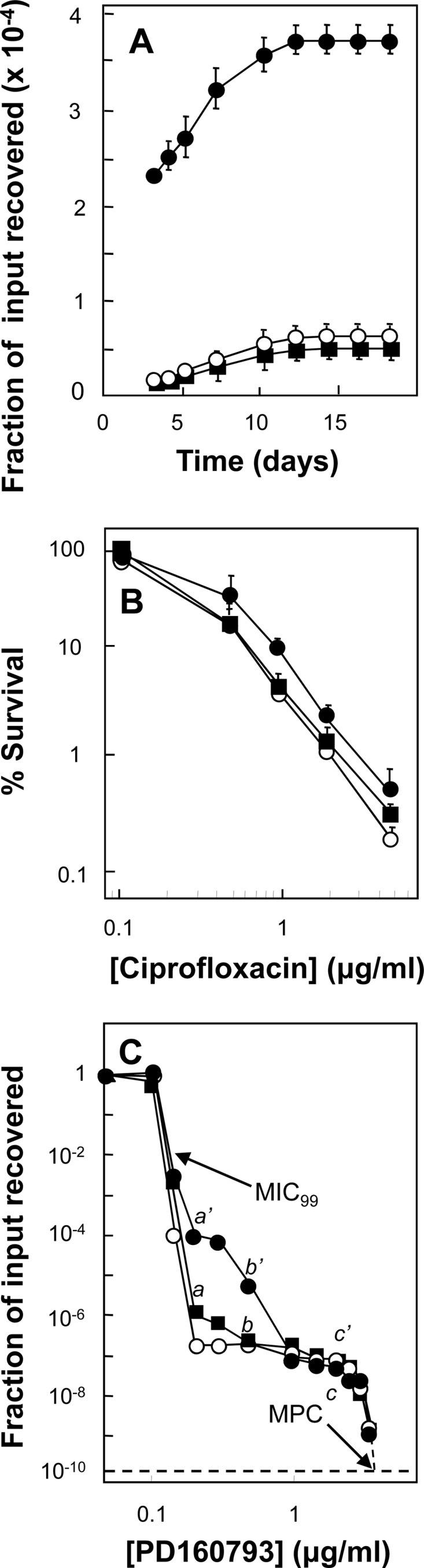
Characterization of a mutator mutation of M. smegmatis. (A) Induction of ciprofloxacin-resistant mutants. M. smegmatis strains were grown to exponential phase, and 3.5 × 106 CFU were applied to agar containing 0.5 μg/ml ciprofloxacin (2.5 times the MIC). At daily intervals, the cumulative number of mutants was recorded and expressed as a fraction of the number of input cells. Symbols: wild type (KD1163), open circles; InhR Rifr mutator (KD2859), filled circles; InhR Rifr mutant obtained by stepwise selection (KD2860), filled squares. Error bars represent standard deviations; replicate experiments gave similar results. (B) Lethal susceptibility to ciprofloxacin. M. smegmatis was incubated at the indicated concentrations of ciprofloxacin for 18 h, and then samples were diluted, applied to agar, and incubated and CFU were counted. Symbols represent the same strains as in panel A. Error bars represent standard deviations; replicate experiments gave similar results. (C) Population analysis with PD160793. Stationary-phase cultures were applied to agar plates containing the indicated concentrations of ciprofloxacin and incubated for 3 days. Colonies were counted and confirmed to be resistant mutants by regrowth on agar containing the same concentration of ciprofloxacin that was initially used for selection. Similar results were obtained in replicate experiments. The symbols are the same as in panel A. The MIC99 and MPC are indicated by arrows. The characteristics of the mutants recovered at points a, b, c, a′, b′, and c′ are listed in Table 3. The dashed line indicates the recovery of 1 mutant per 1010 cells applied to agar, a threshold often used to determine the MPC.
