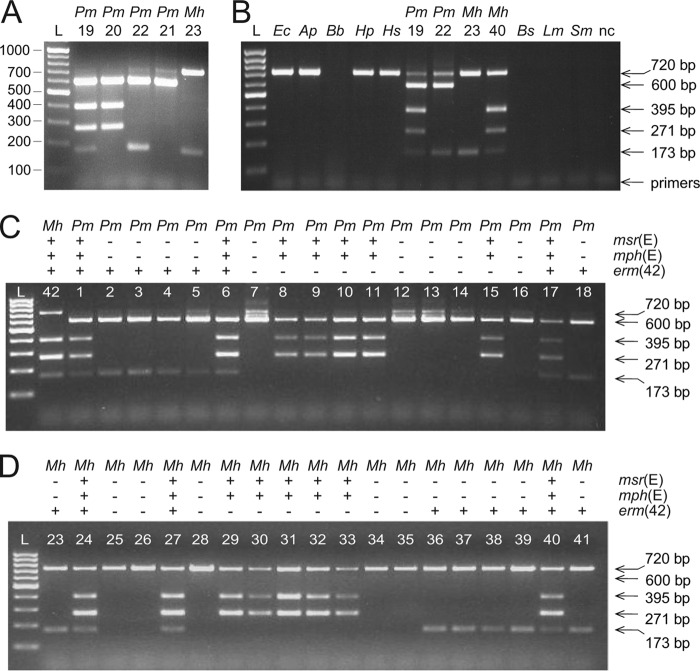
Fig 1.
Gene identification using the multiplex PCR system. (A) The multiplex PCR system was tested on DNA purified from four previously characterized P. multocida (Pm) strains (lanes 19 to 22) and a newly isolated M. haemolytica (Mh) strain (lane 23). PCR fragment sizes can be seen against the 100-bp/step GeneRuler DNA ladder (lane L). (B) Multiplex PCR applied directly to colonies of E. coli (Ec), Actinobacillus pleuropneumoniae (Ap), Bordetella bronchiseptica (Bb), Haemophilus parasuis (Hp), Histophilus somni (Hs), P. multocida, M. haemolytica, Bacillus subtilis (Bs), Listeria monocytogenes (Lm) and Streptococcus mutans (Sm). Templates that gave no products with the multiplex PCR were tested and shown to function in control reactions with species-specific primers (not shown). The 600-bp fragment, which here verifies the presence of P. multocida, is also generated from the DNA of another Pasteurellaceae member, Haemophilus influenzae (not shown). nc, negative control without DNA template. (C) Reactions performed directly on colonies of P. multocida (Pm) strains. None of the strains had been previously genetically characterized. A single M. haemolytica (Mh) strain is included to show species differentiation by formation of the 720-bp, but not the 600-bp, fragment. (D) Reactions performed directly on colonies of M. haemolytica (Mh) strains, none of which had previously been characterized genetically. Species classification and the presence (+) or absence (−) of the erm(42), msr(E), and mph(E) genes are tabulated above the gel lanes. Strain annotations and MIC profiles are given in Table 2.