Fig 1.
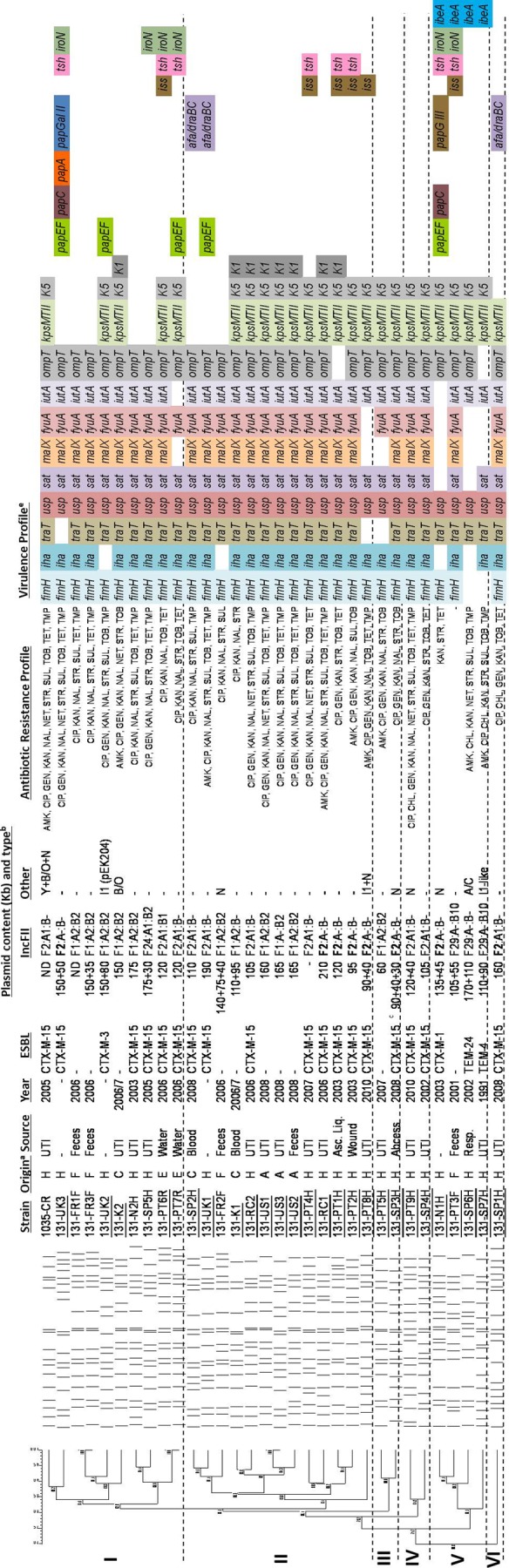
XbaI profiles were analyzed by InfoQuest FP version 5.4 software (Bio-Rad Laboratories), and the percentage similarity was calculated by applying the unweighted-pair group method using average linkages (UPGMA) algorithm based on the Dice coefficient (1.0% band tolerance; 1.0% optimization). (a) Isolates considered ExPEC are underlined. (b) H, hospitalized humans; C, community-acquired infections; F, healthy humans; A, animal; E, environment (marine waters). (c) Plasmids encoding the ESBLs identified in each isolate are shown in bold. FII plasmids were identified using the FAB formula (FII, FIA, FIB), as proposed previously (31). Antibiotic susceptibility testing was done by Etest and the disk diffusion method following CLSI guidelines (3) (d) This isolate coproduces SHV-12. Asc. Liq., ascistic liquid. (e) fimH, type 1 fimbriae; papA, P fimbriae major subunit, pyelonephritis associated; papC, P fimbriae assembly; papEF, P fimbriae minor tip pilins; papG allele II, papG variant, pyelonephritis associated; papG allele III, P fimbriae adhesin, cystitis associated; afa/draBC, Dr antigen-specific adhesin; iha, iron-regulated gene homologue adhesin; sat, secreted autotransporter toxin; tsh, serine protease autotransporter; fyuA, yersiniabactin receptor; iutA, ferric aerobactin receptor; iroN, catecholate siderophore receptor; kpsMTII, group II capsular polysaccharide; kpsMTII K1, variant K1; kpsMTII K5, variant K5; traT, serum survival associated; iss, increased serum survival; ibeA, invasion of brain endothelium; usp, uropathogen-specific protein; ompT, outer membrane protease; malX, pathogenicity-associated island marker. PT, Portugal; SP, Spain; UK, United Kingdom; US, United States of America; FR, France; NW, Norway; RC, Czech Republic; KO, South Korea; SW, Switzerland; CR, Croatia.
