Abstract
Streptococcus pyogenes (group A streptococcus [GAS]) is a highly virulent Gram-positive bacterium. For successful infection, GAS expresses many virulence factors, which are clustered together with transcriptional regulators in distinct genomic regions. Ralp3 is a central regulator of the ERES region. In this study, we investigated the role of Ralp3 in GAS M49 pathogenesis. The inactivation of Ralp3 resulted in reduced attachment to and internalization into human keratinocytes. The Δralp3 mutant failed to survive in human blood and serum, and the hyaluronic acid capsule was slightly decreased. In addition, the mutant showed a lower binding capacity to human plasminogen, and the SpeB activity was significantly decreased. Complementation of the Δralp3 mutant restored the wild-type phenotype. The transcriptome and quantitative reverse transcription-PCR analysis of the serotype M49 GAS strain and its isogenic Δralp3 mutant identified 16 genes as upregulated, and 43 genes were found to be downregulated. Among the downregulated genes, there were open reading frames encoding proteins involved in metabolism (e.g., both lac operons and the fru operon), genes encoding lantibiotics (e.g., the putative salivaricin operon), and ORFs encoding virulence factors (such as the whole Mga core regulon and further genes under Mga control). In summary, the ERES region regulator Ralp3 is an important serotype-specific transcriptional regulator for virulence and metabolic control.
INTRODUCTION
Group A streptococcus (GAS) is a causative agent of human diseases ranging from commonly mild superficial infections of the skin and mucous membranes of the nasopharynx to severe but rare toxic and invasive diseases (2, 5, 7). GAS strains are equipped with an arsenal of virulence factors which allow this pathogen to infect and survive in the human host. Regulation of virulence factor expression is fine-tuned by two-component systems and stand-alone regulators (11, 18). Many GAS virulence factor- and transcriptional regulator-encoding genes cluster together in discrete genomic regions (11). The longest known and best-characterized virulence regulator is Mga, the central regulator of the virulence factor-encoding Mga region (14). This regulator controls genes encoding proteins involved in adherence, internalization, and host immune evasion. Mga also influences the expression of many genes and operons involved in metabolism and sugar utilization (14).
Several studies showed that Mga also interacts with RofA-like protein family (RALP) regulators RofA and Nra, the central regulators of the FCT virulence region (1, 5, 16, 29). Transcriptome analysis of a serotype M49 GAS strain and its isogenic Nra knockout mutant revealed the transcriptional control of another RALP family regulator, Ralp3 (RofA-like protein regulator type 3 [19]). ralp3 homologous genes were exclusively found in serotypes M1, M4, M12, M28, and M49. ralp3 is linked with a gene encoding Epf (extracellular protein factor from Streptococcus suis), a putative plasminogen-binding protein in GAS. Plasminogen acquisition from human host promotes GAS invasion of human keratinocytes via plasminogen-integrin interaction (39). In a serotype-specific fashion, this ralp3 epf gene block is integrated into the eno sagA gene block encoding a plasminogen binding enolase and a streptolysin S precursor. The complete eno ralp3 epf sagA region was thus called the ERES pathogenicity island and is transcriptionally regulated by Ralp3 (19).
The acquisition of different energy sources, the ability to thrive under limited carbon source levels, and the limitation of the transcription of virulence relevant and carbohydrate utilization genes in response to changes in environmental conditions by GAS are absolutely essential for the survival of the bacteria in a human host (24). GAS, as one of the major pathogenic lactic acid bacteria, utilizes glucose as a main carbon source (40). GAS glycolysis has recently been studied by a systems biology approach, and the first GAS glycolytic kinetic model has been published (23).
Different transcriptomic studies have shown that virulence regulation by GAS is linked to genes that are important for sugar utilization (24, 25, 36, 38). In particular, starch-degrading and carbohydrate-metabolism genes are differentially regulated during GAS stationary-phase survival in saliva (37). An extensive remodeling of the transcriptome, including a shutdown of glycolysis and an activation of amino acid catabolism, was observed during Streptococcus pyogenes survival in blood (12). Metabolic adaptations also occurred during pharyngitis in a macaque animal infection model (41) and during mouse soft tissue infections (12).
To characterize the functional and virulence-associated role of Ralp3 encoded in the ERES region of the GAS serotype M49 strain, a ralp3 deletion mutant (19) and its corresponding complemented version was used in several pathogenesis-related assays and microarray analyses.
MATERIALS AND METHODS
Bacterial strains, eukaryotic cells, and culture conditions.
GAS serotype M49 strain 591 was obtained from R. Lütticken (Aachen, Germany). The construction of the Δralp3 mutant and the Δralp3::ralp3 complemented mutant strain has been published by Kreikemeyer et al. (19). The GAS wild-type strain and the mutants were cultured in Todd-Hewitt broth (Invitrogen) supplemented with 0.5% yeast extract (THY; Invitrogen) at 37°C under a 5% CO2 to 20% O2 atmosphere. For selection of the mutants, antibiotics were added to the media at following concentrations: kanamycin, 300 μg ml−1; and erythromycin, 5 μg ml−1.
The human keratinocyte cell line HaCaT (DKFZ, Heidelberg, Germany) was maintained in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 1% glutamine and 10% (vol/vol) fetal bovine serum (Gibco) in tissue culture flasks (Greiner) at 37°C under a 5% CO2 to 20% O2 atmosphere (3).
Eukaryotic cell adherence and internalization and viability/cytotoxicity-assay.
Adherence to and internalization into HaCaT cells was quantified by using an antibiotic protection assay (27). Briefly, 24-well plates were inoculated with 2.5 × 105 cells per well in DMEM without antibiotics. The cells were allowed to grow to confluence. For the assay, cells were washed with DMEM, and infected with GAS M49 strain 591 at a multiplicity of infection (MOI) of 1:10 in DMEM. At 2 h postinfection, the cells were washed extensively with phosphate-buffered saline (PBS), detached from the wells by trypsin treatment, and lysed with sterile distilled water. The viable counts of GAS released from the lysed cells were determined by serial dilution in PBS and plating on THY agar. For the assessment of bacterial internalization, cells were washed at 2 h postinfection with PBS and incubated with DMEM supplemented with penicillin (50 U ml−1) and streptomycin (50 μg ml−1) for an additional 2 h. Subsequently, the cells were washed and lysed, and the GAS viable counts were determined as described above.
To determine the viability of HaCaT cells in the presence of GAS strains, the Live/Dead stain for mammalian cells (Molecular Probes) was used according to the manufacturer's instructions. Bacteria were added to the eukaryotic cells at an MOI of 1:10. The viability of cells was determined after 2, 3, and 4 h of infection using a SpectraMax M2 reader (Molecular Devices).
Capsular hyaluronic acid measurement.
The amount of hyaluronic acid produced by each GAS strain was determined by releasing capsule from exponential-phase GAS cells grown in THY and measuring the hyaluronic acid content of the cell extracts using Stains-All (Sigma) as described previously (35). The absorbance values were compared to a standard curve generated using known concentrations of hyaluronic acid from Streptococcus equi. The amount of hyaluronic acid capsule produced by the tested strains was expressed as femtograms per CFU.
GAS survival in different human media.
The survival assay was performed as described by Nakata et al. (26). Briefly, overnight cultures of the wild-type and mutant strains were inoculated into fresh medium and grown to exponential growth phase. Bacteria were harvested by centrifugation and suspended and diluted in PBS. A 20-μl portion of each suspension was inoculated into 480 μl of heparinized blood and blood serum to a bacterial count of 5 × 103 to 10 × 103 CFU/ml. After 3 h of incubation at 37°C with rotation, the CFU were determined by plating and compared to the inoculum.
Cell surface plasminogen acquisition.
An assay with intact bacteria was performed by using a method adapted from Ringdahl et al. (32). Overnight cultures grown in THY medium were washed in PBS and suspended in one-tenth of the original volume. Then, 100 μl of cells, equivalent to ∼108 CFU, was added to a mixture of 4 ml of PBS supplemented with plasminogen (2 μg ml−1) or PBS (negative control), followed by incubation at 37°C for 1 h. The reactions were terminated by centrifugation at 4°C. The bacterial pellets were washed in ice-cold PBS containing 0.1% Tween 20 and placed on ice. The plasmin activity associated with the bacteria was measured using a method adapted from Kulisek et al. (21). Each pellet was incubated in 100 μl of plasmin substrate solution, prepared by mixing 2 volumes of chromogenic substrate H-d-Val-Leu-Lys-ρ-nitroanilide (Sigma) stock solution (5 mg ml−1) with 3 volumes of 32 mM Tris–1.77 M NaCl (pH 7.5) for 15 min at 37°C, followed by an absorbance measurement at 405 nm.
Cysteine protease activity assay.
The SpeB protease activity in GAS culture supernatants was determined using the method adapted from Hytonen et al. (15). Briefly, the bacteria were grown in THY medium to the transition phase and harvested by centrifugation, and the supernatants were collected. Then, 100 μl of the filtered (0.22-μm pore size) supernatants was incubated with dithiothreitol (DTT; 1 mM) for 30 min at 37°C to activate the enzyme. Next, n-benzoyl-proline-phenylalanine-arginine-p-nitroanilide hydrochloride (1 mM) and phosphate buffer (pH 6; 5 mM) were added to the activated supernatants. The change in absorbance at 405 nm was detected in a spectrophotometer. E64-cystein protease inhibitor was tested in parallel assays to confirm the specificity of the test.
Substrate utilization assay.
The assay was performed as described by Fiedler et al. (9) with small modifications. Briefly, the bacteria were grown overnight in THY, pelleted by centrifugation, washed twice in PBS, and suspended in glucose-free chemical defined medium (CDM [40]). The optical densities (ODs) were adjusted to 0.05, and 100 μl of the bacterial suspension was applied to each well of Biolog Phenotype microarray plates PM1 and PM2. The microarray plates were incubated for 24 h at 37°C in a 5% CO2 and 20% O2 atmosphere, and the OD at 600 nm (OD600) of each well was measured. The ODs in well A1 of the arrays containing no carbon source were subtracted from all values. The ODs in the wells containing α-d-glucose were set to 1, and all other values were correlated accordingly.
For the concentration series carbon source experiments, bacteria were inoculated into 10-ml cultures of CDM supplemented with the indicated carbon sources at different concentrations. The OD600 was measured in hourly intervals.
For the direct competition experiments of wild-type and mutant strain equal CFU of each strain were inoculated into CDM-lactose and CDM-fructose. At time points 0, 4, and 8 h postinoculation, the CFU values for both strains were determined from the mixtures by plating serial dilutions on agar plates. The antibiotic resistance of the mutant allowed a numerical discrimination and evaluation. Wild-type and mutant single cultures grown under the same conditions served as controls.
RNA preparation, microarray analysis, and quantitative reverse transcription-PCR (qRT-PCR).
Total RNA was isolated from GAS serotype M49 and its isogenic ralp3 mutant in eight independent cultures grown to the transition phase (OD600 = 1.0) with FastRNA Pro Blue Kit (MP Biomedicals). cDNA synthesis was performed with 10 μg of total RNA. The RNA was mixed with random primers (Promega) and spike-ins (Two-Color RNA Spike-In Kit; Agilent Technologies), followed by incubation at 70°C for 10 min and 5 min on ice, respectively. Then, the following reagents were added: 10 μl of 5× First Strand Buffer (Invitrogen), 5 μl of 0.1 M DTT (Invitrogen), 0.5 μl of a deoxynucleoside triphosphate mix (10 mM dATP, dGTP, and dTTP, 2.5 mM dCTP), 1.25 μl of Cy3-dCTP or Cy5-dCTP (GE Healthcare), and 2 μl of the SuperScript II reverse transcriptase (Invitrogen). The mixture was incubated at 42°C for 60 min and then heated to 70°C for 10 min, followed by incubation on ice for 5 min. The RNA was degraded by incubation with 2 U of RNase H (Invitrogen) at room temperature for 30 min. Labeled cDNA was purified using a CyScribe GFX purification kit (GE Healthcare). A total of 200 ng of Cy3-labeled cDNA and 200 ng of Cy5-labeled cDNA were hybridized together to the microarray (Custom Array GE 8x15K; Agilent). Microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE30397 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30397).
For qRT-PCR the synthesis of cDNA was performed using the Superscript first-strand synthesis system for RT-PCR (Invitrogen). Primer sets for selected genes are listed in Table 1. The real-time PCR amplification was performed with SYBR green (Fermentas) using an ABI Prism 7000 sequence detection system (Applied Biosystems). The level of DNA gyrase subunit A gene (gyrA) transcription was used for normalization. For data comparison, the normalized values were calculated by log2 expression ratio.
Table 1.
qRT-PCR primers used in this study
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| fruR | fruR-M49-for | ATGGCAAAGATCACTGAAGAAAACT |
| fruR-M49-rev | TTCTTGCTCCAACTCTCCTAGAT | |
| fruK | fruK-M49-for | CGTGACCTTAAACCCTTCTATTG |
| fruK-M49-rev | CCTCCAGCAAACTTATCATCACT | |
| lacA | lacA-M49-for | TTAGGCCATTTTATTGAGCATGTCA |
| lacA-M49-rev | TCCAGAATTAGCGAAGAATATCGTC | |
| salT | salT-M49-for | TCATTTTACCTCTTCCTCCTGATAT |
| salT-M49-rev | TGACTATGGAAACCATCATTATTTGT | |
| salA | salA-M49-for | TTAACAACAAACAAATACTGAGTTCG |
| salA-M49-rev | TGAAAAAGAACTTATGGAAGTAGCTG | |
| lacE | lacE-M49-for | TTAATTTTCAAACTGTGCTTTTACAAAATC |
| lacE-M49-rev | TGATATTAAATTAGCTAAAACAGAAGGG | |
| lacA2 | lacA2-M49-for | TTAGCACATTTTATTAAGCATGTCGAC |
| lacA2-M49-rev | TCCAGGAATTGCAAAGAACATTATC | |
| emm49 | emm49-for | AACGCTAAAGTAGCCCCACAAG |
| emm49-rev | TCGCCTGTTGACGGTAACG | |
| mga | mga-M49-for | TTCACAGATAACAACGTTATGGTCAA |
| mga-M49-rev | TTCTGGTTTTTGTACCTCTTTGTCACT | |
| sof | sof-M49-for | CAG TTC GAT TAT ACC AAG CGT GTA G |
| sof-M49-rev | TGAGGGCTACTACTACCACAGTTATTCAAAGCGA | |
| malP | malP-M49-for | ATGGAAACAATAAGCAAGAAAATGCC |
| malP-M49-rev | GATAAACAGGCAAGGGAACGG | |
| copY | copY-M49-for | TTAATACGTCTCCTTTTTCTGACAG |
| copY-M49-rev | TTGCCAATTGCTTGAAAGGAAAAAAC | |
| hrcA | hrcA-M49-for | TCAGTGAACTTCATAATGGTTAGAATTT |
| hrcA-M49-rev | TAGGTCTGTTAGTTTGGTAAACATTATT | |
| scrR | scrR-M49-for | GTGGTCGCAAAATTAACAGACGT |
| scrR-M49-rev | TGTGTTTTGGGAAAGGTAGCCTT | |
| gyrA | gyrA-M49-for | CGACTTGTCTGACGCCAAA |
| gyrA-M49-rev | TTATCACGTTCCAAACCAGTCAA |
Statistical analysis.
The significance of differences between samples in plasminogen binding, adherence, internalization, viability/cytotoxicity assays, capsular hyaluronic measurement, and survival in different media was determined using the two-tailed U test. P < 0.05 was defined as marginally significant. P < 0.01 was defined as significant and P < 0.001 was defined as highly significant. The results are depicted as averages with the standard deviation (n ≥ 5). The significance of differences between samples in substrate utilization assays was determined using two-tailed Student t test. The results are shown as averages and the standard deviation (n = 3).
RESULTS
Requirement of Ralp3 for GAS adherence and internalization.
Since adherence to and internalization into host epithelial cells are essential steps for successful infection by GAS, we assayed the impact of Ralp3 on the adherence to and internalization into human keratinocytes. The GAS serotype M49 strain was isolated from skin infection, so we decided to use keratinocytes as a model for adherence and internalization by GAS M49. As shown in Fig. 1A and B, after 2 h of infection the adherence to and internalization of the Δralp3 mutant into HaCaT cells was significantly reduced. Complementation of the Δralp3 mutant by gene expression from a plasmid restored wild-type keratinocyte adherence and internalization levels. In addition to the adherence and internalization assay, the viability of eukaryotic cells was determined after 2, 3, and 4 h of infection. No significant differences between wild-type and mutant strains were detected (Fig. 1C).
Fig 1.
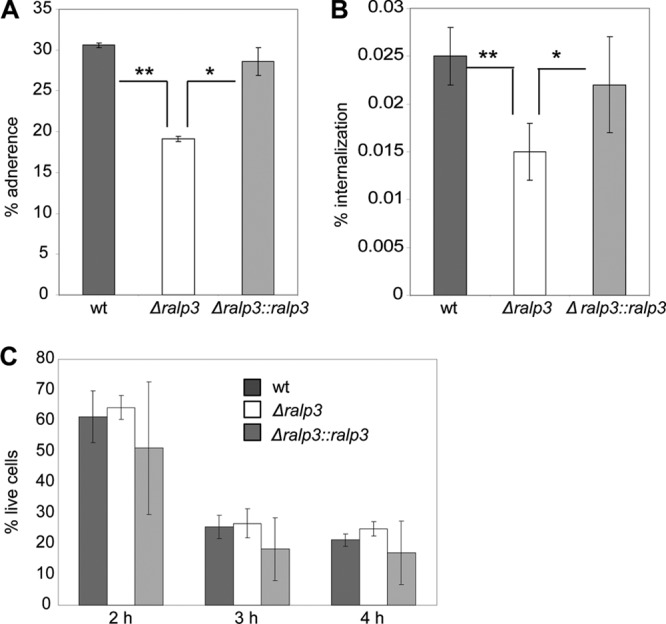
Adherence to (A) and internalization into (B) human keratinocytes by GAS M49 wild-type, Δralp3, and Δralp3::ralp3 mutant strains. (C) Cytotoxic effect of wild-type and mutant strains on human keratinocytes. The data represent mean values ± the standard deviations from three independent experiments. Only significant differences (*, P < 0.05; **, P < 0.01) are indicated by asterisks.
GAS survival in different media.
Next, we tested the growth behavior of all strains in THY medium. No changes were detected between wild-type and mutant strains (Fig. 2A). Subsequently, we determined the ability of wild-type and mutant strains to survive in different human media as described by Nakata et al. (26). As demonstrated in Fig. 2B and C, the Δralp3 mutant was strongly attenuated in its ability to survive in human blood and serum. The multiplication factor was <1.0. These results indicate that Ralp3 regulatory activity substantially contributes to evasion of the host immune response in the blood environment.
Fig 2.
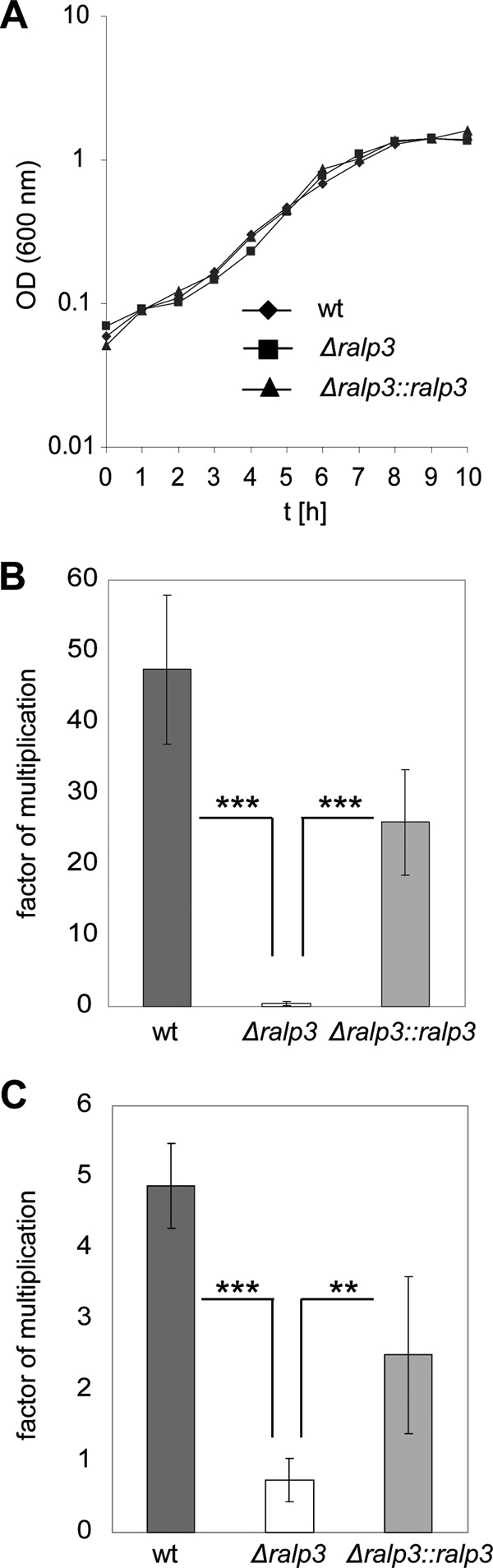
Growth of wild-type and mutant strains in THY medium (A) and survival in human whole blood (B) and human serum (C). The data of the survival assays represent mean values ± the standard deviations from three independent experiments. The number of surviving CFU was determined by plating serial dilutions and subsequent counting. The y axis shows the resulting multiplication factor for each strain calculated from the percentage of surviving CFU related to the inoculum. Only significant differences (**, P < 0.01; ***, P < 0.001) are indicated by asterisks.
The hyaluronic acid capsule is decreased in the Δralp3 mutant.
Capsule expression is strongly linked to survival of GAS in human blood (42). Kwinn et al. described Ralp3 as a transcriptional regulator of hasA in GAS serotype M1 (22). hasA encodes a hyaluronate synthase involved in capsule formation, which has a known role in resistance to phagocytic clearance (42). To determine the amount of hyaluronic acid capsule produced by each GAS strain, cells from exponential-phase cultures were used. As shown in Fig. 3A, the hyaluronic acid capsule of the Δralp3 mutant is slightly reduced. The level of hyaluronic acid in the mutant strain was 3.0 fg/CFU lower compared to the wild-type strain. Whether this observation explains any of the survival phenotypes needs to be investigated in greater detail.
Fig 3.
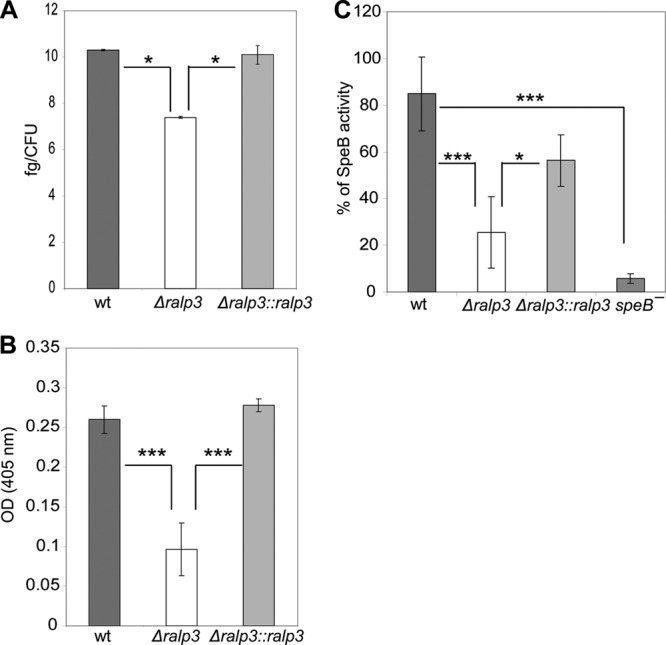
(A) Capsular hyaluronic acid measurement of wild-type and mutant strains. Absorbance values were compared to a standard curve generated using known concentrations of hyaluronic acid. The y axis shows the resulting concentrations (in fg/CFU) of hyaluronic acid for each strain calculated from concentrations related to the inoculum CFU. (B) Plasminogen acquisition by wild-type and mutant strains. The data represent mean values ± the standard deviations from five independent experiments. The y axis shows the OD405 for each strain. (C) SpeB activity of culture supernatants. The supernatants were activated by DTT, and the proteolytic cleavage of the substrate was measured at OD405. Only significant differences (*, P < 0.05; ***, P < 0.001) are indicated by asterisks.
Plasminogen acquisition is modulated by Ralp3.
Our previous study suggested that Epf is a potential GAS plasminogen-binding protein, and Ralp3 acts as a positive regulator of epf transcription. The binding of plasminogen by Epf was previously demonstrated by indirect adherence enzyme-linked immunosorbent assays with recombinant Epf (19). We tried to confirm the control of epf by Ralp3 using plasminogen acquisition assays with intact bacteria. As demonstrated in Fig. 3B, the acquisition of plasminogen was significantly reduced by up to 63% in the Δralp3 mutant. The reverse complementation of the Δralp3 mutant strain by ralp3 from the GAS M49 wild type restored the ability to acquire plasminogen.
SpeB activity is decreased in Δralp3 culture supernatants.
GAS is able to secrete enzymatically active cysteine protease SpeB. We compared the SpeB activity of the GAS M49 wild-type strain to that of the Δralp3 and Δralp3::ralp3 mutant strains. A ΔspeB mutant was used as a negative control. The SpeB activity of the wild type was set 100%, and the measurements of the supernatants of the mutants were related to the wild-type stain. As shown in Fig. 3C, the Δralp3 mutant showed significantly decreased SpeB activity.
Δralp3 transcriptome analysis.
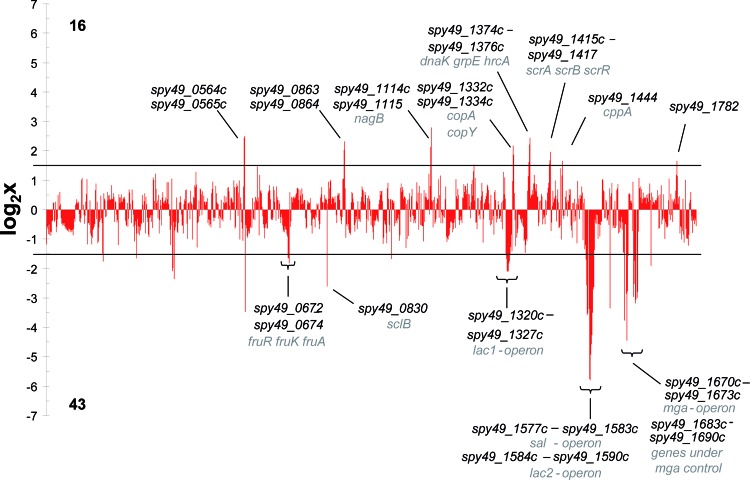
The previous series of experiments documented a quite complex regulatory influence of Ralp3 on GAS M49 virulence phenotypes. To characterize the Ralp3 regulatory function in GAS for the first time on the whole-genome level, GAS M49 wild-type and Δralp3 mutant strains were comprehensively compared by two-color microarray analysis. Samples were taken from cultures in the transition phase because the transcriptional maximum of ralp3 has been reported in this phase (19). Figure 4 highlights the differences between the wild-type and Δralp3 mutant strains. Accordingly, the transcription of 16 genes (1.0%) in the Δralp3 mutant was found to be upregulated. In addition, 43 genes (2.5%) were found to be considerably downregulated in the Δralp3 mutant. The majority of genes (59.4%) did not show significant differences and further 37.1% of the genes showed no active transcripts in the medium and growth phase tested.
Fig 4.
Overview of the transcript levels in the Δralp3 mutant in the transition phase. Log2 expression ratios of the mutant are shown. All genes with log values >1.58 are significantly induced, and genes with a negative log of <−1.58 are significantly reduced. All genes between the lines are considered as not significantly influenced.
Genes differentially expressed in the Δralp3 mutant.
The detailed results for the 59 open reading frames (ORFs) with a ≥3.0-fold change in their transcript levels in the Δralp3 mutant strain compared to the wild type are listed in Table 2. The transcript level profile was determined by log2 expression ratio. Genes were considered significantly different when the log2 ratio of the fold change was ≤−1.58 or ≥1.58. Among the downregulated genes, there were genes encoding for proteins involved in metabolic processes, e.g., both lac operons (lacABC1D1 and lacA2B2C2D2EFG) and the fru operon (fruAKR). Furthermore, the genes of the putative salivaricin operon (salABTXYK), which encodes for a putative lantibiotic, were downregulated in the mutant background. Apart from genes that are involved in metabolic processes, there were several virulence factor genes with decreased transcript levels. The whole Mga core regulon (mga, spy49_1672, emm49, ennX, and spy49_1668) and genes under Mga control (sfbX49, sof, and speB) showed significantly decreased amounts of transcripts.
Table 2.
Genes down- and upregulated by ralp3 knockout in S. pyogenes M49 (strain 591)a
| ORF | Gene | Description | Log2 expression ratio |
||||
|---|---|---|---|---|---|---|---|
| Array 1 | Array 2 | Avg | SD | qRT-PCRb | |||
| Downregulated genes | |||||||
| spy49_0162 | opuABC | l-Proline glycine betaine ABC transport system permease | −1.75 | −1.69 | −1.72 | 0.05 | NT |
| spy49_0254c | Transposase | −1.65 | −1.71 | −1.68 | 0.04 | NT | |
| spy49_0343 | Hypothetical protein | −2.09 | −2.04 | −2.07 | 0.03 | NT | |
| spy49_0344 | Hypothetical protein | −2.00 | −1.97 | −1.99 | 0.02 | NT | |
| spy49_0347 | Hypothetical protein | −2.36 | −2.40 | −2.38 | 0.03 | NT | |
| spy49_0672 | fruR | Putative transcriptional repressor | −1.64 | −1.64 | −1.64 | 0.00 | −1.61 ± 0.88 |
| spy49_0673 | fruK | Tagatose-6-phosphate kinase/1-phosphofructokinase | −1.65 | −1.69 | −1.67 | 0.03 | −1.80 ± 0.98 |
| spy49_0674 | fruA | PTS system, fructose-specific IIABC component | −1.82 | −1.87 | −1.84 | 0.04 | NT |
| spy49_0830 | sclB | SclB protein | −2.60 | −2.64 | −2.62 | 0.02 | NT |
| spy49_1024 | Hypothetical protein | −1.68 | −1.71 | −1.69 | 0.02 | NT | |
| spy49_1321c | Hypothetical protein | −1.68 | −1.72 | −1.70 | 0.03 | NT | |
| spy49_1322c | lacD1 | Tagatose 1,6-diphosphate aldolase | −2.08 | −2.07 | −2.07 | 0.01 | NT |
| spy49_1323c | lacC1 | Tagatose-6-phosphate kinase/1- phosphofructokinase | −2.10 | −2.08 | −2.09 | 0.01 | NT |
| spy49_1324c | Tagatose-6-phosphate kinase/1- phosphofructokinase | −2.09 | −2.08 | −2.09 | 0.00 | NT | |
| spy49_1325c | lacB | Galactose-6-phosphate isomerase subunit LacB | −1.81 | −1.80 | −1.81 | 0.01 | NT |
| spy49_1326c | lacA | Galactose-6-phosphate isomerase subunit LacA | −1.73 | −1.76 | −1.75 | 0.02 | −1.97 ± 0.75 |
| spy49_1577c | salK | SalK | −1.71 | −1.67 | −1.69 | 0.02 | NT |
| spy49_1578c | salY | Putative ABC transporter | −2.57 | −2.69 | −2.63 | 0.08 | NT |
| spy49_1579c | salX | Putative salivaricin A ABC transporter | −3.58 | −3.95 | −3.77 | 0.26 | NT |
| spy49_1580c | salT | Putative salivaricin A modification enzyme | −3.52 | −4.58 | −4.05 | 0.75 | −4.21 ± 1.08 |
| spy49_1582c | salB | Putative salivaricin A modification enzyme, amino acid dehydration | −3.37 | −4.57 | −3.97 | 0.85 | NT |
| spy49_1583c | salA | Lantibiotic salivaricin A precursor | −5.75 | −5.71 | −5.73 | 0.03 | −6.01 ± 0.45 |
| spy49_1584c | lacG | 6-Phospho-β-galactosidase | −5.80 | −5.88 | −5.84 | 0.06 | NT |
| spy49_1585c | lacE | PTS system, lactose-specific IIBC component (EIIBC-Lac) | −4.94 | −4.98 | −4.96 | 0.03 | −5.45 ± 1.08 |
| spy49_1586c | lacF | PTS system, lactose-specific IIA component (EIIA-Lac) | −4.59 | −4.56 | −4.57 | 0.02 | NT |
| spy49_1587c | lacD2 | Tagatose 1,6-diphosphate aldolase | −4.26 | −4.42 | −4.34 | 0.12 | NT |
| spy49_1588c | lacC2 | Tagatose-6-phosphate kinase | −3.50 | −3.64 | −3.57 | 0.10 | NT |
| spy49_1589c | lacB2 | Galactose-6-phosphate isomerase subunit LacB | −2.70 | −2.66 | −2.68 | 0.03 | NT |
| spy49_1590c | lacA2 | Galactose-6-phosphate isomerase subunit LacA | −2.62 | −2.66 | −2.64 | 0.03 | −1.61 ± 2.08 |
| spy49_1633c | sclA | Collagen-like surface protein | −3.35 | −3.39 | −3.37 | 0.03 | NT |
| spy49_1668c | Cell surface/fibronectin-binding protein | −3.77 | −3.89 | −3.83 | 0.08 | NT | |
| spy49_1670c | ennX | EnnX protein | −2.86 | −2.86 | −2.86 | 0.00 | NT |
| spy49_1671c | emm49 | Antiphagocytic M protein | −4.45 | −4.51 | −4.48 | 0.04 | −5.16 ± 0.65 |
| spy49_1672c | Fc-gamma receptor | −2.79 | −2.76 | −2.77 | 0.02 | NT | |
| spy49_1673c | mga | M protein trans-acting positive regulator (Mga) | −1.58 | −1.81 | −1.70 | 0.17 | −1.36 ± 0.16 |
| spy49_1683c | sfbX49 | Fibronectin-binding protein | −2.97 | −3.06 | −3.01 | 0.06 | NT |
| spy49_1684c | sof | Serum opacity factor | −2.54 | −3.03 | −2.78 | 0.35 | −2.71 ± 0.90 |
| spy49_1686 | Hypothetical protein | −1.73 | −2.30 | −2.02 | 0.40 | NT | |
| spy49_1687c | ropA | Proteinase maturation protein (RopA) | −3.18 | −3.15 | −3.16 | 0.02 | NT |
| spy49_1688c | Hypothetical protein | −3.07 | −3.08 | −3.08 | 0.01 | NT | |
| spy49_1689c | Spi SpeB protease inhibitor | −3.00 | −3.09 | −3.04 | 0.06 | NT | |
| spy49_1690c | speB | Streptococcal pyrogenic exotoxin B (SpeB) | −2.79 | −2.87 | −2.83 | 0.06 | NT |
| spy49_1719c | csp | Major cold-shock protein | −1.90 | −1.80 | −1.85 | 0.07 | NT |
| Upregulated genes | |||||||
| spy49_0564c | Transposase | 2.45 | 2.47 | 2.46 | 0.01 | NT | |
| spy49_0565c | Transposase | 2.51 | 2.43 | 2.47 | 0.06 | NT | |
| spy49_0863 | malP | Putative l-malate permease | 2.03 | 2.06 | 2.05 | 0.02 | 1.13 ± 0.50 |
| spy49_0864 | NAD-dependent malic enzyme | 2.33 | 2.25 | 2.29 | 0.06 | NT | |
| spy49_1114c | Putative 16S pseudouridylate synthetase | 2.13 | 2.26 | 2.20 | 0.09 | NT | |
| spy49_1115c | nagB | Glucosamine-6-phosphate deaminase | 2.80 | 2.97 | 2.88 | 0.12 | NT |
| spy49_1239 | Alpha-mannosidase | 1.53 | 1.63 | 1.58 | 0.07 | NT | |
| spy49_1332c | copA | Putative copper-transporting ATPase | 2.18 | 2.18 | 2.18 | 0.00 | NT |
| spy49_1334c | copY | Negative transcriptional regulator, CopY | 1.86 | 1.87 | 1.87 | 0.01 | 2.41 ± 0.12 |
| spy49_1374c | dnaK | Molecular chaperone DnaK | 1.61 | 1.50 | 1.56 | 0.08 | NT |
| spy49_1375c | grpE | Heat shock protein GrpE | 2.21 | 2.25 | 2.23 | 0.03 | NT |
| spy49_1376c | hrcA | Heat-inducible transcription repressor | 2.44 | 2.47 | 2.45 | 0.02 | 3.05 ± 0.39 |
| spy49_1416 | scrB | Sucrose-6-phosphate hydrolase | 1.67 | 2.00 | 1.83 | 0.23 | NT |
| spy49_1417 | scrR | Putative sucrose operon repressor | 1.96 | 1.96 | 1.96 | 0.00 | 1.48 ± 0.62 |
| spy49_1444c | cppA | Putative putative C3-degrading proteinase | 1.65 | 1.63 | 1.64 | 0.02 | NT |
| spy49_1782 | Hypothetical protein | 1.65 | 1.71 | 1.68 | 0.04 | NT | |
Genes are listed according to the genomic location of the ORFs annotated in the GAS serotype M49 strain NZ131 and are considered differentially expressed if the log2 of the fold change was ≤−1.58 or ≥1.58. Data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE30397.
Only significant values are shown (P ≤ 0.05). NT, not tested.
Among the genes with increased transcript abundance in the Δralp3 mutant were ORFs encoding for proteins involved in metabolism (malP, scrB, and scrR), degradation (cppA), and stress response (dnaK, grpE, and hrcA). In the case of the downregulated genes, there was a preference observed for genes clustered in operon/regulon structures. This bias was not found for the upregulated genes in the Δralp3 mutant.
To confirm the microarray data, transcript levels of several down- and upregulated genes were determined by qRT-PCR. Consistent with the microarray data, all 14 tested genes showed transcript level changes in the same direction (Table 2).
Substrate utilization by GAS M49 wild type and Δralp3 mutant.
Surprisingly, the microarray data revealed the downregulation of genes encoding proteins with metabolic function, especially ORFs responsible for sugar utilization. To determine the effect of ralp3 deletion on the ability of GAS M49 to utilize different carbon sources, Biolog phenotype microarrays were used. A total of 190 different carbon sources were tested with these microarrays. Growth at 10% (= 0.1) on α-d-glucose was defined as vital growth. Both the wild-type and Δralp3 mutant strains were able to utilize 20 carbon sources. These were N-acetyl-d-glucoseamine, tween40, tween80, d-fructose, α-d-glucose, α-d-lactose, d-trehalose, maltose, maltotriose, d-mannose, sucrose, salicin, d-glucoseamine, pectin, uridine, mannan, N-acetyl-β-d-mannosamine, β-methyl-d-glucoside, dextrin, and gelatin. Exclusively, the wild-type strain was able to utilize 3-O-β-d-galactopyranosyl-d-arabinose. The Δralp3 mutant showed an increased growth on d-lactose, d-trehalose, and maltotriose. Significant differences in the utilization of carbon sources are listed in Table 3.
Table 3.
GAS M49 and its isogenic ralp3 mutant were analyzed for their ability to utilize different carbon sourcesa
| Substrate | GAS M49 (mean OD ± SD) |
||
|---|---|---|---|
| Wild type | Δralp3 mutant | P | |
| α-d-Lactose | 0.29 ± 0.06 | 0.77 ± 0.18 | 0.032 |
| d-Trehalose | 0.47 ± 0.03 | 0.73 ± 0.05 | 0.001 |
| Maltose | 0.87 ± 0.05 | 0.71 ± 0.05 | 0.041 |
| Maltotriose | 0.77 ± 0.06 | 1.16 ± 0.09 | 0.005 |
| 3-O-β-d-Galactopyranosyl-d-arabinose | 0.27 ± 0.02 | 0.04 ± 0.06 | 0.005 |
Only significant differences between wild-type and mutant strains are listed. The optical densities (ODs) of the wild-type culture grown on α-d-glucose were set to 1, and the ODs of growth of wild-type and Δralp3 mutant strains on other carbon sources are related to this value.
In addition, growth experiments in batch cultures with different amounts of sugars (CDM supplemented with glucose, maltose, fructose, or lactose) were performed. As shown in Fig. S1 in the supplemental material, no differences in growth behavior between wild-type and mutant strains were observed in CDM-glucose and CDM-maltose. Again, the Δralp3 deletion mutant reached higher ODs at all tested concentrations of lactose. On CDM-fructose, slight differences between the wild-type and mutant strains were noted in the exponential-growth phase, suggesting either a delayed uptake or metabolism of fructose in the mutant. However, both strains reached the same final OD at the stationary growth phase.
In order to analyze whether the wild type or Δralp3 deletion mutant has any selective growth advantage in direct competition, we performed growth experiments with both strains at equal starting CFU in the same culture. As shown in Fig. S2 in the supplemental material, at the 0-h time point postinoculation equal numbers of CFU were recovered from the mixed cultures, thereby confirming equal starting levels of CFU. No differences in CFU recovery between the strains were observed at different growth stages on CDM-lactose. During growth on CDM-fructose, the wild-type strain achieved a higher CFU number at the exponential (4 h) and stationary (8 h) growth phases. Apparently, the wild-type strain suppressed the growth of the Δralp3 deletion mutant or at least outcompeted the mutant to a significant level.
DISCUSSION
In the present study we investigate the impact of the deletion of the Ralp3 regulator on GAS M49 virulence factor expression. The RALP family regulators were characterized in four variants. RofA, also titled Ralp1, and Nra, also designated Ralp2, are encoded in a serotype-specific fashion within the FCT genomic region (1, 16, 18, 29). Analysis of the GAS serotype M1 genome sequence (8) led to the identification of two additional Ralp regulators, Ralp3 and Ralp4 (13). Both were partially characterized (19, 22). Our analyses indicate that Ralp3 is a transcriptional regulator involved in GAS virulence, thereby confirming the general observation from Kwinn et al. (22). However, the Ralp3 regulatory directions are apparently GAS serotype specific, as noted in the previous study (19) and as confirmed by results shown here. The comparison of the data from 2007 (22) and the present study is summarized in Fig. S3 in the supplemental material.
One of the initial steps of infection by GAS is the attachment to host surfaces and the internalization into these. These processes have been well characterized in recent years (6, 18). Our results show that the transcriptional regulator Ralp3 is involved in the control of host cell adherence processes. The Δralp3 mutant showed reduced adherence to and internalization into human keratinocytes. The microarray data support these results. The transcript levels of the Mga regulon genes are decreased. In particular, genes encoding for fibronectin-binding proteins (spy49_1668c and sfbX49), a collagen-like protein (sclA), and the antiphagocytic M-protein (emm49) are significantly reduced. The role of fibronectin-binding proteins and the Mga regulon in GAS attachment and internalization is well characterized (10, 16, 17, 20). The repression of the transcription of these genes leads to a reduced number of these important surface-associated proteins and results in reduced attachment to eukaryotic structures.
Kwinn et al. (22) described Ralp3 in a serotype M1T1 as a direct or indirect transcriptional repressor for capsule synthesis. The synthesis of the hyaluronic acid capsule (42) is an important strategy to circumvent the host immune response and enables GAS to survive in human blood or serum. In GAS M49 the knockout of ralp3 impairs the ability of the bacteria to multiply in human media. One explanation for this phenotype could be a moderately reduced hyaluronic acid capsule in the Δralp3 mutant. However, no corresponding change in hasA transcript was noted. Most likely, changes in emm gene transcription and subsequent M protein amounts are more important for these observations. The transcriptome data confirm these results. The expression of emm49, encoding for antiphagocytic M protein, and the expression of the whole sal operon is reduced. SalY is required for full virulence and intracellular survival in macrophages (28). The reduced transcript amounts of both genes leads to a decreased protection against phagocytosis and impaired survival in human media. In addition, hasA is regulated by Mga (30), but the expression of mga is reduced in the Δralp3 mutant strain. Therefore, our results indicate that in contrast to M1 in the M49 serotype, Ralp3 could act as a transcriptional inducer of hasA.
A surprising result of the Ralp3 transition-phase whole transcriptome presented here is the apparent lack of correspondence to our previously published Ralp3 regulation data, which were based on luciferase gene reporter strains (19). Apart from the divergent methodological approaches, the reason for this fact is the definition of the cutoff. Genes from the present study were considered as significantly different when the log2 ratio of the fold change was ≤−1.58 or ≥1.58. Based on the luciferase reporter gene assays of the previous study (19), genes that showed different transcription in the Δralp3 deletion mutant over the complete growth phase were generally defined as regulated. Reevaluation and calculation of the previous single gene luciferase reporter gene assay data and comparison with the data sets for the same genes from the DNA array experiments described here revealed that only two values are not consistent (shown in Table 4). The gene encoding Nra, which is located outside of the ERES region, seems to be downregulated in our previous study (19), if regulation criteria from the present study were applied. The gene encoding Epf from the ERES region showed no signal in the array experiments, whereas decreased transcript levels were deduced from the luciferase reporter gene assay in the previous study (19). All other tested genes showed no different transcript levels using the regulation criteria of the present study. The reason for some of these discrepancies could be the different sensitivities and specificities of the assays used in both studies. The transcription of the luciferase-encoding gene depends on the promoter structure of each gene under investigation, subsequent translation efficiency, and finally the activity of the enzyme could be also attenuated or induced.
Table 4.
Comparison of the transcriptomic data from a 2007 report (Kreikemeyer et al. [19]) and the present study for selected genes
| Gene | Log2 expression ratio as determined in: |
|
|---|---|---|
| 2007 | 2012 | |
| nra | −3.32 | −1.38 |
| cpa | −1.81 | −1.24 |
| msmR | 0.46 | 2.02 |
| prtF2 | 1.38 | 2.12 |
| eno | 0.14 | 1.69 |
| epf | −2 | − |
| sagA | 0.58 | −1.47 |
An important observation made here was that Ralp3 also controls gene transcription from operons encoding sugar utilization enzymes. These include both lac operons and the fru operon, which are well characterized in different Gram-positive bacteria (4, 33, 34, 43). GAS encodes two lactose metabolic operons. The lac2 operon is involved in carbohydrate metabolism and encodes genes required in the tagatose-6-phosphate pathway (33). However, the lac1 operon seems to be involved in virulence gene regulation (25). Furthermore, Mga influences directly or indirectly over 10% of the GAS transcriptome, including genes encoded for sugar utilization, e.g., the fru operon (31). The knockout of ralp3 resulted in a reduction in the transcript levels of Mga, which in turn could lead to reduced transcription of the fru operon. The question remains why some previously reported transcriptional effects of Mga mutation are not exactly mirrored in the Δralp3 mutant transcriptome, which also revealed a downregulation of mga. Direct versus indirect regulatory effects, as well as thus-far-unknown regulator hierarchies, are possible explanations. These questions need to be addressed in future studies.
By screening 190 different carbon sources, we found differences in only five sugars. Exclusively the wild-type strain was able to utilize 3-O-β-d-galactopyranosyl-d-arabinose. The utilization of maltose was significantly increased in this strain. Surprisingly, the Δralp3 mutant grew on α-d-lactose much better than did the wild type, although the lac operon was suppressed in the mutant. The same results were observed in batch culture experiments. No differences were detected during the growth in CDM-glucose and -maltose. In CDM-fructose, the Δralp3 deletion mutant showed slower growth in the exponential phase. This could be the reason for the observed phenotype in the direct competition experiment. The wild-type and mutant strains were inoculated at the same CFU. After entering the exponential growth phase, the wild-type strain obviously utilized the fructose more efficiently, resulting in a significant outcompetition of the mutant growth. Batch culture experiments in CDM-lactose confirmed the observed phenotype from microarray experiments. The Δralp3 deletion mutant reached higher ODs. No differences in direct competition in CDM with lactose as the sole carbon source were observed. The reason for this phenotype remains unclear and is currently being investigated in our lab by probing genome-scale and kinetic models of carbohydrate metabolism in a GAS M49 genomic background.
It should be noted that further in vitro studies in different carbohydrate sources, also in direct competition setup, most likely will not lead to meaningful results. The regulative consequences that the Δralp3 deletion exerts on carbohydrate metabolism in this mutant need to be investigated in an appropriate animal infection model. Such studies are under way.
In summary, we have shown that Ralp3 is an important regulator, which connects virulence regulation with metabolic activity. Complete deletion of ralp3 negatively affects the transcription of virulence and metabolic genes, resulting in reduced virulence and survival characteristics and changes in sugar utilization. Whether Ralp3 control of its target genes is direct or indirect is under investigation.
The question of whether Ralp3 as a part of the RE gene block is exclusively present in certain GAS serotypes and whether there is a serotype-specific regulatory network needs to be evaluated in future studies in order to acquire a deeper understanding of GAS pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
The work in the lab of B.K. was supported by the following grants awarded from the Federal Ministry of Education and Research (BMBF): BMBF ERANet “Pathogenomics” II and BMBF ERANet “SysMO-System Biology of Microorganism” II. We further acknowledge receiving funds from the Rostock University Medical Faculty FORUN program.
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bessen DE, Kalia A. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bisno AL, Brito MO, Collins CM. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191–200 [DOI] [PubMed] [Google Scholar]
- 3. Boukamp P, et al. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burne RA, Wen ZT, Chen YY, Penders JE. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685–694 [DOI] [PubMed] [Google Scholar]
- 6. Courtney HS, Hasty DL, Dale JB. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 34:77–87 [DOI] [PubMed] [Google Scholar]
- 7. Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferretti JJ, et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fiedler T, et al. 2011. Characterization of three lactic acid bacteria and their isogenic ldh deletion mutants shows optimization for YATP (cell mass produced per mole of ATP) at their physiological pHs. Appl. Environ. Microbiol. 77:612–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fiedler T, et al. 2010. Impact of the Streptococcus pyogenes Mga regulator on human matrix protein binding and interaction with eukaryotic cells. Int. J. Med. Microbiol. 300:248–258 [DOI] [PubMed] [Google Scholar]
- 11. Fiedler T, Sugareva V, Patenge N, Kreikemeyer B. 2010. Insights into Streptococcus pyogenes pathogenesis from transcriptome studies. Future Microbiol. 5:1675–1694 [DOI] [PubMed] [Google Scholar]
- 12. Graham MR, et al. 2006. Analysis of the transcriptome of group A streptococcus in mouse soft tissue infection. Am. J. Pathol. 169:927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Granok AB, Parsonage D, Ross RP, Caparon MG. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hondorp ER, McIver KS. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 66:1056–1065 [DOI] [PubMed] [Google Scholar]
- 15. Hytonen J, Haataja S, Gerlach D, Podbielski A, Finne J. 2001. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding, and cysteine protease activity. Mol. Microbiol. 39:512–519 [DOI] [PubMed] [Google Scholar]
- 16. Kreikemeyer B, Klenk M, Podbielski A. 2004. The intracellular status of Streptococcus pyogenes: role of extracellular matrix-binding proteins and their regulation. Int. J. Med. Microbiol. 294:177–188 [DOI] [PubMed] [Google Scholar]
- 17. Kreikemeyer B, Martin DR, Chhatwal GS. 1999. SfbII protein, a fibronectin binding surface protein of group A streptococci, is a serum opacity factor with high serotype-specific apolipoproteinase activity. FEMS Microbiol. Lett. 178:305–311 [DOI] [PubMed] [Google Scholar]
- 18. Kreikemeyer B, McIver KS, Podbielski A. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224–232 [DOI] [PubMed] [Google Scholar]
- 19. Kreikemeyer B, et al. 2007. The Streptococcus pyogenes serotype M49 Nra-Ralp3 transcriptional regulatory network and its control of virulence factor expression from the novel eno ralp3 epf sagA pathogenicity region. Infect. Immun. 75:5698–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kreikemeyer B, Oehmcke S, Nakata M, Hoffrogge R, Podbielski A. 2004. Streptococcus pyogenes fibronectin-binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279:15850–15859 [DOI] [PubMed] [Google Scholar]
- 21. Kulisek ES, Holm SE, Johnston KH. 1989. A chromogenic assay for the detection of plasmin generated by plasminogen activator immobilized on nitrocellulose using a para-nitroanilide synthetic peptide substrate. Anal. Biochem. 177:78–84 [DOI] [PubMed] [Google Scholar]
- 22. Kwinn LA, et al. 2007. Genetic characterization and virulence role of the RALP3/LSA locus upstream of the streptolysin s operon in invasive M1T1 group A streptococcus. J. Bacteriol. 189:1322–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levering J, et al. 2012. Role of phosphate in the central metabolism of two lactic acid bacteria: a comparative systems biology approach. FEBS J. 279:1274–1290 [DOI] [PubMed] [Google Scholar]
- 24. Loughman JA, Caparon MG. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 25:5414–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loughman JA, Caparon MG. 2007. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64:269–280 [DOI] [PubMed] [Google Scholar]
- 26. Nakata M, et al. 2009. Mode of expression and functional characterization of FCT-3 pilus region-encoded proteins in Streptococcus pyogenes serotype M49. Infect. Immun. 77:32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ozeri V, Rosenshine I, Mosher DF, Fassler R, Hanski E. 1998. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 30:625–637 [DOI] [PubMed] [Google Scholar]
- 28. Phelps HA, Neely MN. 2007. SalY of the Streptococcus pyogenes lantibiotic locus is required for full virulence and intracellular survival in macrophages. Infect. Immun. 75:4541–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Podbielski A, Woischnik M, Leonard BA, Schmidt KH. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051–1064 [DOI] [PubMed] [Google Scholar]
- 30. Podbielski A, Woischnik M, Pohl B, Schmidt KH. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. 185:171–181 [DOI] [PubMed] [Google Scholar]
- 31. Ribardo DA, McIver KS. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62:491–508 [DOI] [PubMed] [Google Scholar]
- 32. Ringdahl U, et al. 1998. Molecular cooperation between protein PAM and streptokinase for plasmin acquisition by Streptococcus pyogenes. J. Biol. Chem. 273:6424–6430 [DOI] [PubMed] [Google Scholar]
- 33. Rosey EL, Oskouian B, Stewart GC. 1991. Lactose metabolism by Staphylococcus aureus: characterization of lacABCD, the structural genes of the tagatose 6-phosphate pathway. J. Bacteriol. 173:5992–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosey EL, Stewart GC. 1992. Nucleotide and deduced amino acid sequences of the lacR, lacABCD, and lacFE genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon of Streptococcus mutans. J. Bacteriol. 174:6159–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schrager HM, Rheinwald JG, Wessels MR. 1996. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J. Clin. Invest. 98:1954–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shelburne SA, et al. 2007. MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins. J. Bacteriol. 189:2610–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shelburne SA, et al. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 105:1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shelburne SA, et al. 2007. Regulation of polysaccharide utilization contributes to the persistence of group a streptococcus in the oropharynx. Infect. Immun. 75:2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siemens N, Patenge N, Otto J, Fiedler T, Kreikemeyer B. 2011. Streptococcus pyogenes M49 plasminogen/plasmin binding facilitates keratinocyte invasion via integrin-integrin-linked kinase (ILK) pathways and protects from macrophage killing. J. Biol. Chem. 286:21612–21622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Virtaneva K, et al. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. U. S. A. 102:9014–9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc. Natl. Acad. Sci. U. S. A. 88:8317–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J. Bacteriol. 192:2434–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.