Fig 1.
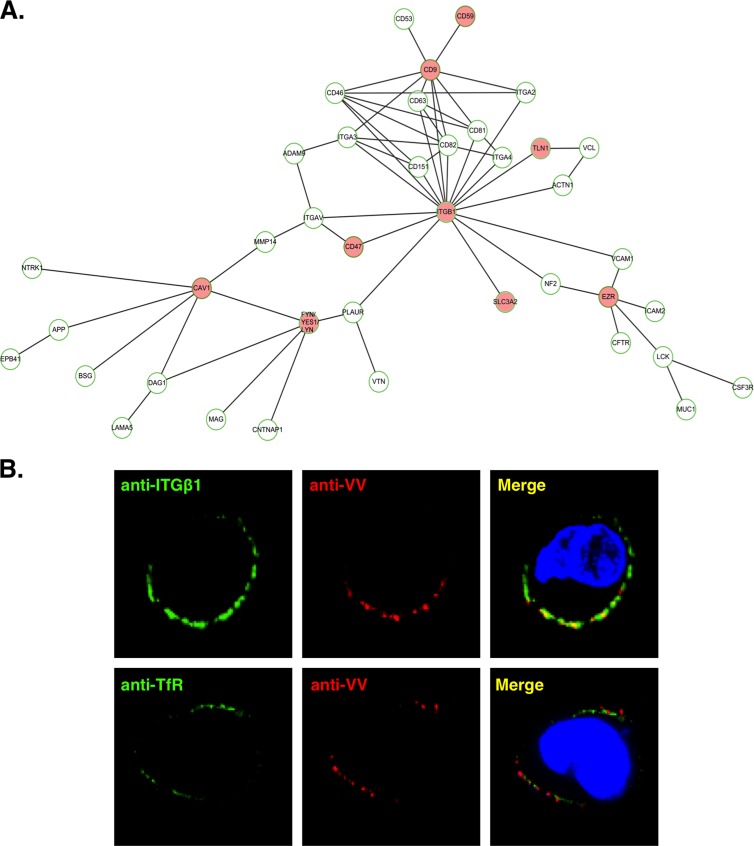
Integrin β1 was identified in lipid rafts and is associated with vaccinia MV on HeLa cells. (A) The hierarchical ITGβ1 network, constructed by ARIADNE Pathway Studio 7.0 software (see Materials and Methods), shows integrin β1 and its direct and indirect interacting membrane proteins. The nodes are labeled with gene symbols, and the red nodes represent the cellular proteins identified in the lipid raft fractions upon vaccinia MV infections (55). ITGβ1, integrin β1; TLN1, talin-1; EZR, ezrin; CAV1, caveolin-1; SLC3A2, CD98. (B) Vaccinia MV colocalizes with ITGβ1 at the surface of HeLa cells. HeLa cells were infected with vaccinia MV at an MOI of 50 PFU per cell for 1 h at 4°C, washed, and transferred to 12°C, where the cells were incubated with rabbit anti-VV antibody (1:500) (red) and anti-integrin β1 MAb (12G10) (1:1,000) (green) for 1 h for copatching as previously described (30). Alternatively, anti-VV antibody was incubated with anti-transferrin receptor (TfR) (green) MAb as a control. Tetramethylrhodamine-conjugated goat anti-rabbit IgG (1:1,000) and FITC-conjugated goat anti-mouse IgG (1:1,000) were subsequently added, and cells were fixed for confocal microscopy. DNA was visualized by 4′-6-diamidino-2-phenylindole (DAPI) staining (blue).