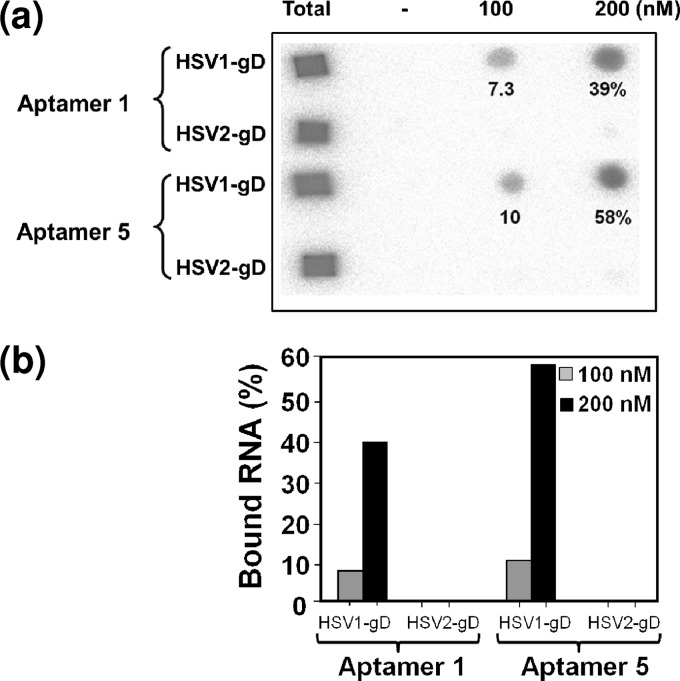
Fig 3.
Filter binding analyses of the selected aptamers. (a) Filter binding assays were performed with unmodified RNA (2′OH) in the presence of gD proteins (100 and 200 nM) from HSV-1 and HSV-2, and tRNA was used as a nonspecific competitor. (b) The amounts of labeled aptamers (unmodified) retained on the filter in the presence and absence of gD proteins were quantitated with an image analyzer (BAS2000; Fuji Film). The percentages of binding to the gD protein of HSV-1 were plotted.